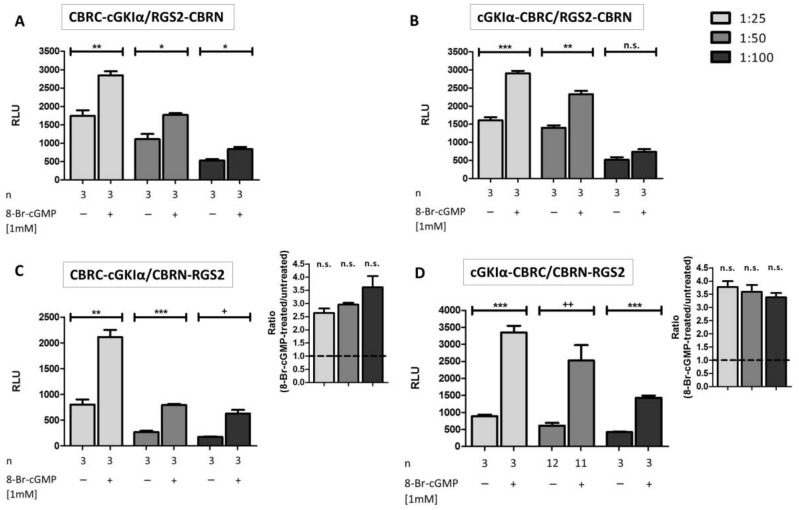
Figure 3.
Influence of vector ratio for transfection and subsequent interaction analysis of cGKIα and RGS2: COS7-cells were seeded in 6-well plates (3.3 × 105 cells/well) and co-transfected with 4 different combinations of cGKIα/RGS2-vectors (each transfection with 15 µg DNA, vector ratio as indicated). After transfer to 96-well plates (1.0 × 104 cells/well) and addition of 1 mM 8-Br-cGMP, cells were incubated for 24 h. In most cases, a significant signal-increase was observed (A–D). The most promising vector combinations for enhanced luminescence upon 8-Br-cGMP application were CBRC-cGKIα/CBRN-RGS2 (C) and cGKIα-CBRC/CBRN-RGS2 (D); ratiometric analysis (8-Br-cGMP-treated/untreated) can be seen on the inset of (C,D), respectively. Data is expressed as mean ± standard error of the mean (SEM). For unpaired Student’s t-test p-values < 0.05 were considered significant (*, +), <0.01 and <0.001 highly significant (**, ++ and ***, respectively), whereas statistical differences characterized by an + were calculated by unpaired Student’s t-test with Welch’s correction. A non-significant difference was marked as n.s. N = technical replicates. RLU: relative luminescence unit.