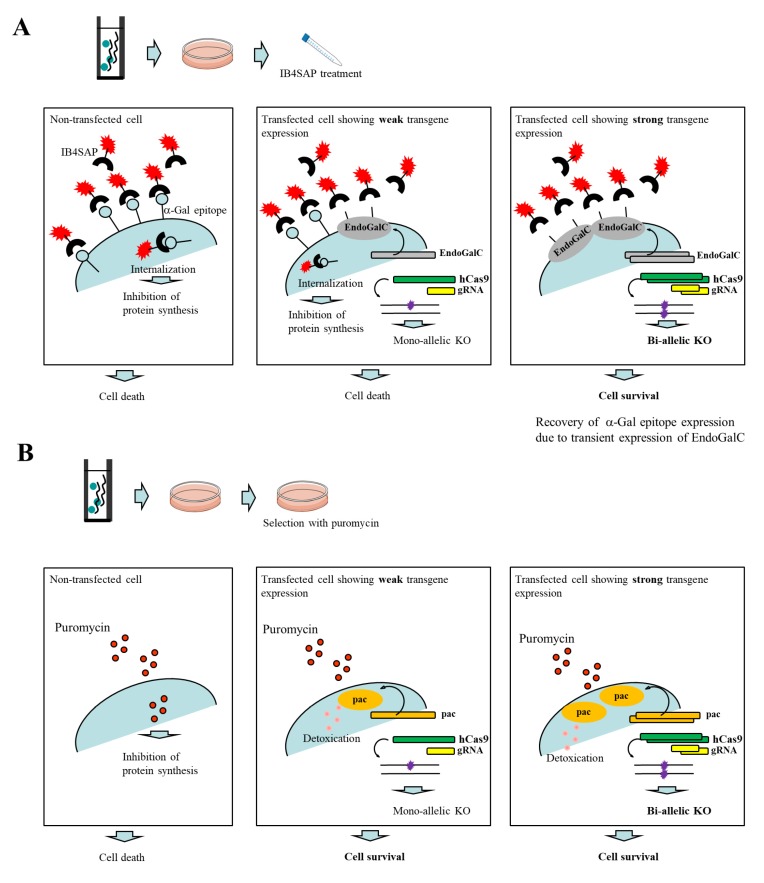
Figure 1.
(A) Schematic representation of the isolation of genome-edited cells using EndoGalC-mediated digestion of the α-Gal epitope, CRISPR/Cas9-based genome editing, and IB4SAP-mediated targeted toxin technology. When cells are transfected with three expression vectors containing EndoGalC, hCas9, or gRNA targeted to a target locus, and 3 days later they are treated with IB4SAP for a short period, cells strongly expressing EndoGalC will survive and concomitantly both alleles of a target locus are expected to be knocked out (shown in the right panel). In contrast, cells weakly expressing EndoGalC and untransfected cells (all of which still express the α-Gal epitope on their surface), will undergo cell death (shown in the middle panel). (B) Schematic representation of the isolation of genome-edited cells using conventional puromycin-based genome editing. When cells are transfected with three expression vectors containing pac, hCas9, or gRNA targeted to a target locus, and 2~3 days later they are selected with puromycin for 2 days, the surviving clones are expected to be a mixture of those with mono-allelic knockout (KO) and bi-allelic KO phenotypes (shown in the middle and right panels).