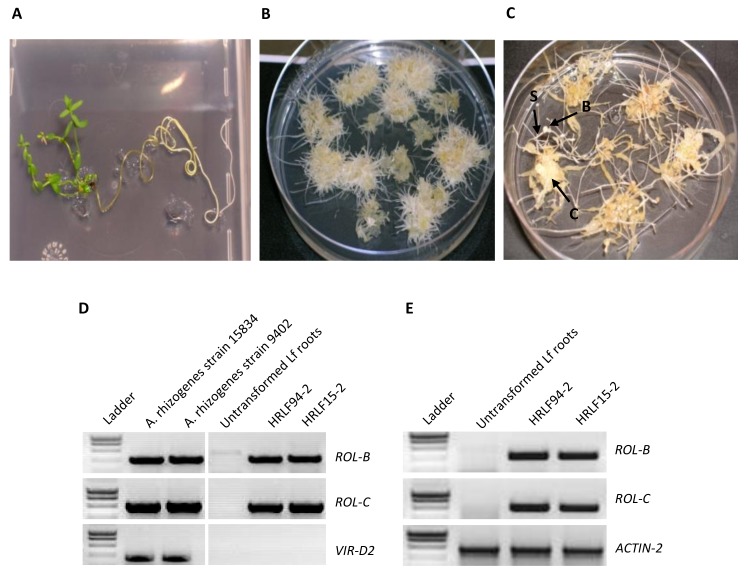
Figure 2.
A. rhizogenes-mediated transformation and molecular characterization of the resulting L. flavum HR lines. (A) Germination of wild type Linum flavum; (B) Hairy roots of Linum flavum, line HRLF15.2; (C) Hairy roots of Linum flavum, line HRLF94-6 (C: callus, B: bud, S: shoot); (D) PCR amplified DNA fragments of ROL-B, ROL-C and VIR-D2 pro-oncogenes from the plasmidic DNA of A. rhizogenes strains or genomic DNA of L. flavum. From left to right: Ladder, DNA from the plasmids of A. rhizogenes LBA 9402 and ATCC 15834 strains respectively, used as positive control, genomic DNA of wild type roots L. flavum cultivated in vitro used as negative control, genomic DNA from two transgenic L. flavum hairy roots lines, HRLF94-2 and HRLF15-2 respectively. (E) sqRT-PCR analysis of ROL-B and ROL-C pro-oncogene expressions in L. flavum wild type roots and in two transgenic L. flavum hairy roots lines, HRLF94-2 and HRLF15-2 respectively. Total RNAs isolated from hairy roots were subjected to sqRT-PCR analysis using ACTIN-2 gene as internal control. Ten microliters of RT-PCR products were loaded on 1% (w/v) agarose gel.