Abstract
Domestication and breeding for human‐desired morphological traits can reduce population genetic diversity via founder events and artificial selection, resulting in inbreeding depression and genetic disorders. The ferret (Mustela putorius furo) was domesticated from European polecats (M. putorius), transported to multiple continents, and has been artificially selected for several traits. The ferret is now a common pet, a laboratory model organism, and feral ferrets can impact native biodiversity. We hypothesized global ferret trade resulted in distinct international genetic clusters and that ferrets transported to other continents would have lower genetic diversity than ferrets from Europe because of extreme founder events and no hybridization with wild polecats or genetically diverse ferrets. To assess these hypotheses, we genotyped 765 ferrets at 31 microsatellites from 11 countries among the continents of North America, Europe, and Australia and estimated population structure and genetic diversity. Fifteen M. putorius were genotyped for comparison. Our study indicated ferrets exhibit geographically distinct clusters and highlights the low genetic variation in certain countries. Australian and North American clusters have the lowest genetic diversities and highest inbreeding metrics whereas the United Kingdom (UK) cluster exhibited intermediate genetic diversity. Non‐UK European ferrets had high genetic diversity, possibly a result of introgression with wild polecats. Notably, Hungarian ferrets had the highest genetic diversity and Hungary is the only country sampled with two wild polecat species. Our research has broad social, economic, and biomedical importance. Ferret owners and veterinarians should be made aware of potential inbreeding depression. Breeders in North America and Australia would benefit by incorporating genetically diverse ferrets from mainland Europe. Laboratories using ferrets as biomedical organisms should consider diversifying their genetic stock and incorporating genetic information into bioassays. These results also have forensic applications for conserving the genetics of wild polecat species and for identifying and managing sources of feral ferrets causing ecosystem damage.
Keywords: artificial selection, Australia, Europe, Mustela putorius, Mustela putorius furo, New Zealand, North America
1. INTRODUCTION
Domestication can result in a founder effect where only a few wild animals contribute to the gene pool and evolutionary trajectory of the domesticated lineage (Diamond, 2002; Mimura et al., 2017; Petersson, Jaurvi, Steffner, & Ragnarsson, 1996; Teletchea & Fontaine, 2014). Additionally, artificial selection for desirable morphological traits can further reduce genetic variation (Driscoll, Macdonald, & O'Brien, 2009; Leroy, 2011; Muñoz‐Fuentes et al., 2014). This loss of genetic diversity can result in inbreeding depression (Charlesworth & Charlesworth, 1999; Hedrick & Garcia‐Dorado, 2016; Leroy, 2014), which can be particularly problematic when a domesticated species is economically or ecologically important (González‐Recio, López de Maturana, & Gutiérrez, 2007; O'Neill, Church, McGreevy, Thomson, & Brodbelt, 2013; Rhymer & Simberloff, 1996). Thus, understanding the population structure and genetic diversity of a domestic species is critically important for long‐term persistence and can have global or regional implications based on the animal's role in society and its genetic status.
The ferret (Mustela putorius furo Linnaeus, 1758) was domesticated from the European polecat (M. putorius Linnaeus, 1758) primarily for hunting rabbits and rats (Blandford, 1987; Hosoda et al., 2000; Kurose, Abramov, & Masuda, 2000, 2008). More recently, the ferret has become a common household pet (Hernádi, Kis, Turcsán, & Topál, 2012) and, in some countries, is a laboratory model organism (Ball, 2006) or considered invasive (O'Donnell, Weston, & Monks, 2017). Selection for specific ferret coat colors has been associated with genetically determined physical abnormalities (Blaszczyk et al., 2007; Piazza, Abitbol, Gnirs, Huynh, & Cauzinille, 2014), and there is rising concern about the potential role of inbreeding and low genetic diversity in the increasing incidence of ferret cancers (Avallone et al., 2016; Bielinska, Parviainen, Kiiveri, Heikinheimo, & Wilson, 2009; Clagett, Johnston, & Han, 2016; Lewington, 2007b). Thus, for ferrets, an assessment of population structure and regional genetic diversity will be of broad social, economic, and biomedical importance.
On a global scale, pet owners and veterinarians would be informed of genetic diversity and potential inbreeding depression (Fox & Marini, 2014) and breeders trying to avoid genetic disorders would benefit by applying information about genetic diversity to regional and international breeding programs (Howard, Lynch, Santymire, Marinari, & Wildt, 2015; Willoughby et al., 2015). Biomedical laboratories using ferrets as model organisms would benefit by incorporating genetic diversity information into their bioassays because inbred individuals can be more susceptible to disease exposures (Ball, 2006; Ilmonen et al., 2008). These data could also be used to identify and conserve wild polecat species which could face reductions in genetic diversity when hybridized with feral ferrets (Costa et al., 2013; Rhymer & Simberloff, 1996). Similarly, the conservation genetics approach used here could have forensic and wildlife management applications for identifying and managing sources of feral ferrets causing ecosystem damage (O'Donnell et al., 2017; Wells, 2009).
Despite the worldwide distribution of ferrets, global patterns of domestic ferret population structure and genetic diversity have not been characterized (Costa et al., 2013; Ernest, Drazenovich, Dalbeck, & Hawkins, 2012; Thomson, 1951). Intercontinental translocations from Europe have not been well recorded, and the current understanding of ferret transportation and domestication relies heavily on transgenerational word of mouth (Church, 2007; Lewington, 2007a) and historic letters (Buller, 1877). Patterns of domestication are also clouded by the historic backcrossing with M. putorius (Costa et al., 2013; Davison et al., 1999; Marmi, López‐Giráldez, & Domingo‐Roura, 2004; Pitt, 1921; Poole, 1972) and potential hybridization with other polecat species (Lodé, Guiral, & Peltier, 2005; Williams et al., 1996). Although historical artificial selection for hunting ability (Carnegie, 2013; Owen, 1984) and contemporary artificial selection for coat colors and patterns (Blaszczyk et al., 2007; Lewington, 2007b; Piazza et al., 2014) likely had a large impact on ferret translocations and genetic diversity, international differences in trade laws and breeding programs could also have limited or reduced the genetic diversity of certain populations (Lee, 2002; Northern Territory Government, 2016; Queensland Government, 2016; Willoughby et al., 2015).
Our goal was to evaluate the genetic structure and levels of genetic diversity and inbreeding in pet ferrets from multiple countries among the continents of North America, Europe, and Australia. We hypothesized global ferret trade resulted in distinct international genetic clusters. We also hypothesized ferrets transported to other continents would have lower genetic diversity than ferrets from Europe because of extreme founder events and no opportunities to hybridize with wild polecats or genetically diverse ferrets. Our international and intercontinental assessment of ferret population genetics provides broad insights into global patterns of founder events, and the impacts of isolation and inbreeding on the population structure of a domesticated species (Larson & Burger, 2013). Our results will be of broad social, economic, and biomedical importance and have direct applications to the ferret industry.
2. MATERIALS AND METHODS
2.1. Sample collection and DNA extraction
Cells for DNA extraction were collected from 765 domestic ferrets (M. putorius furo; Australia: 222; Canada: 56; Denmark: 60; England: 63; Hungary: 19; the Netherlands: 48; Norway: 41; New Zealand: 74; Scotland: 16; Sweden: 27; United States: 139) and 15 European polecats (M. putorius; all from England) via buccal swab and/or hair (>10 hairs per ferret) plucked from the base of the tail (Table S1). Samples were collected by coauthors or via collaborating veterinarians between 2008 and 2011 from personal homes, rescue shelters, or breeders. During that time, cells from a single specimen that died in 2007 were collected from an Australian museum. The polecats were sampled from private breeders who had captured the individuals as wild polecats. Most polecats originated near the western border of England, which is an area known to have polecats with a high amount of introgression with ferrets (Costa et al., 2013). Any individuals known (via breeding programs) or suspected by the owners to be hybrids were removed from analyses and were not included in this sample. Swab samples were stored at −70°C, whereas hair was stored in paper envelopes at stable room temperature. DNA was extracted from buccal swabs or hair follicles from 2010 to 2011 following the exact protocols of Ernest et al. (2012).
2.2. PCR
Forward primers for 31 microsatellite loci (Table S2; Dallas & Piertney, 1998; Domingo‐Roura et al., 2003; Ernest et al., 2012; Lam, Gagne, & Ernest, 2016; O'Connell, Wright, & Farid, 1996; Paetkau & Strobeck, 1994) were fluorescently labeled (NED, PET, FAM, or VIC; Applied Biosystems Inc., Foster City, CA, USA). Two loci were amplified singly, and the other 29 loci were split among seven multiplexes based on fragment size and fluorescent label compatibility (Table S2). Amplifications were carried out in Bio‐Rad MyCyclers (Bio‐Rad, Hercules, CA, USA) using Ernest et al. (2012) multiplex PCR protocols (Table S2). PCR products were analyzed on an ABI 3730 capillary DNA Analyzer (Life Technologies, Carlsbad, CA, USA). Negative controls and positive controls were included with each PCR run. Fragments were visualized with STRand version 2.3.69 (Toonen & Hughes, 2001). Heterozygous and homozygous loci were run at least twice or three times, respectively.
2.3. Population genetic structure
We used three approaches to assess population structure, including F statistics, Bayesian population assignment models, and a discriminant analysis of principal components (DAPC). Population divergence (F ST) was calculated in GenAlEx 6.502 (Peakall & Smouse, 2006, 2012), and significance testing was based on 999 permutations. We used spatially explicit hierarchical Bayesian clustering programs GENELAND 4.0 (Guillot, Mortier, & Estoup, 2005) and TESS 2.3 (Durand, Chen, & François, 2009) to assess population assignments, and adegenet 2.0.1 (Jombart, 2008) for the DAPC.
In GENELAND, the number of populations (K) is a parameter optimized by the model. We followed developer recommendations for determining K and individual population assignments (Guillot, Estoup, Mortier, & Cosson, 2005). First, we ran 15 spatial models allowing K to vary from 1 to 10. All models converged on the same K. Thus, we ran five additional models fixing K at the mode and selected the model with the highest log‐likelihood to run an admixture model. Each run included 100,000 iterations, a thinning interval of 1,000, and a 25% burn‐in period prior to extracting model output.
In TESS, K must be specified and tested over a range of possible values. Model selection must be used to determine the K with the best fit to the data. We followed developer instructions for determining K and population assignments. First, we ran 10 nonadmixture models for each K from 2 to 10. For model comparisons, TESS computes a deviance information criterion (DIC). We ran 10 spatially conditional autoregressive admixture models for each K to the DIC plateau of nonadmixture models. All models included pairwise great circle geographic distances for weighting the Voronoi neighborhood, 100,000 iterations, and a 25% burn‐in period. We retained 20% of the models which contained the lowest DIC scores and used CLUMPP 1.1.2 to perform model averaging (Jakobsson & Rosenberg, 2007).
To assess whether spatial priors or sample size were driving population assignments (Puechmaille, 2016), we also used program STRUCTURE 2.3.4 (Pritchard et al., 2000). We randomly subsetted the samples from each country to 15, which was the number of wild polecats sampled. Using 180 subsetted samples, we ran 10 admixture models for each K from 1 to 10 with uniform, nonspatial priors, including 100,000 iterations and a 25% burn‐in period.
Because the algorithm for individual assignments in adegenet is not as powerful as Bayesian population assignment algorithms (Jombart, Devillard, & Balloux, 2010), we defined populations in the DAPC using countries as focal groups and treated wild European polecats as a separate group. We retained all principal components for discriminant analyses.
2.4. Microsatellite loci and genetic diversity
Tests for linkage disequilibrium, deviations from Hardy–Weinberg proportions, and null alleles were assessed in GENEPOP 4.5.1 (Rousset, 2008). Given the major differences in sample sizes among countries, we used genetic diversity estimates that are robust to sample size or accounted for sample size. We calculated unbiased expected heterozygosity in GenAlEx (Nei, 1978). To measure the number of alleles, we calculated allelic richness using sample size‐correcting rarefaction methods in FSTAT 2.9.3.2 (Goudet, 1995; Kalinowski, 2004). To assess inbreeding, we calculated internal relatedness using package Rhh 1.0.2 in Program R 3.3.0 (Alho, Valimaki, & Merila, 2010). Internal relatedness measures a relative outbred–inbred continuum, where negative values are suggestive of outbred individuals and positive scores are suggestive of inbreeding (Amos et al., 2001). Internal relatedness is an extension of standardized heterozygosity which standardizes estimates based on allele frequencies of the entire global sample of ferrets (Amos et al., 2001). We reported both the raw number or private alleles and the percent of private alleles, which was standardized to sample size.
3. RESULTS
3.1. Population genetic structure
All 31 loci were polymorphic among pooled samples. Only 3% of individuals had any missing data. Most individuals with missing data were missing allelic information at a single locus; however, two individuals had two missing loci, and a single individual had four missing loci. There was no evidence for null alleles or deviations from Hardy–Weinberg proportions in each assigned population after Bonferroni corrections. European polecats had the lowest F ST values with ferrets from England and Scotland and the highest differentiation with ferrets from Australia, Canada, and the United States (Table 1). Australia had high pairwise F ST values with every country except New Zealand and the Netherlands. European countries were not strongly differentiated but exhibited moderate differentiation with the United States and Canada. When population differentiation was assessed based on population assignments in GENELAND, the United Kingdom (UK: England & Scotland ferrets) cluster was not strongly differentiated from non‐UK European cluster (Table 1). The North American cluster was strongly differentiated from the Australian cluster and moderately differentiated from both European clusters.
Table 1.
Summary of country‐level pairwise F ST (below axis) and GENELAND genetic cluster‐level pairwise F ST (above axis) estimates
| Country | Genetic cluster | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Australia | – | Non‐UK Europe | Australia | UK & NZ | |||||||
| Canada | 0.09 | – | 0.08 | 0.14 | 0.08 | North America | |||||
| Denmark | 0.08 | 0.04 | – | – | 0.11 | 0.03 | Non‐UK Europe | ||||
| England | 0.08 | 0.05 | 0.03 | – | – | 0.11 | Australia | ||||
| Hungary | 0.07 | 0.06 | 0.03 | 0.05 | – | ||||||
| The Netherlands | 0.06 | 0.06 | 0.03 | 0.04 | 0.03 | – | |||||
| Norway | 0.07 | 0.05 | 0.01 | 0.02 | 0.03 | 0.03 | – | ||||
| NZ | 0.06 | 0.07 | 0.04 | 0.03 | 0.05 | 0.05 | 0.02 | – | |||
| Scotland | 0.09 | 0.07 | 0.03 | 0.01 | 0.05 | 0.04 | 0.02 | 0.04 | – | ||
| Sweden | 0.09 | 0.05 | 0.02 | 0.04 | 0.06 | 0.05 | 0.02 | 0.05 | 0.05 | – | |
| USA | 0.09 | 0.03 | 0.06 | 0.05 | 0.06 | 0.05 | 0.06 | 0.07 | 0.07 | 0.06 | – |
| Polecat | 0.13 | 0.09 | 0.06 | 0.05 | 0.06 | 0.07 | 0.06 | 0.08 | 0.05 | 0.07 | 0.09 |
| Australia | Canada | Denmark | England | Hungary | The Netherlands | Norway | NZ | Scotland | Sweden | USA |
UK, United Kingdom; NZ, New Zealand; USA, United States of America.
All pairwise F ST estimates were significant (p < .05 based on 1,000 permutation tests) except the England–Scotland comparison.
Program GENELAND identified four genetic clusters (Figure 1a), including distinct North American and Australian clusters. Europe had two clusters, separating the UK ferrets from non‐UK European ferrets. New Zealand ferrets primarily assigned to the UK cluster, but also exhibited admixture with the Australian cluster. Norway also had a large proportion of individuals assigned to the UK cluster. Program TESS assigned individuals similarly, but found additional substructure within Canada and within the Netherlands (Figure 1b). European polecats, sampled in England, primarily assigned to the UK cluster in both programs. On a subsetted dataset, with no spatial priors and equal sample size among countries (N = 15), program STRUCTURE also indicated six clusters based on model probability (LnP(D); Figure 1c). The major geographic trends were similar among all programs; however, STRUCTURE assigned the majority of polecats distinctly from ferrets (Figure 1c), indicating spatial priors in GENELAND and TESS may have overridden the genetic differences among polecats and ferrets in the UK. Alternatively, the same results might have been observed if equalization of sample sizes were performed in programs GENELAND and TESS.
Figure 1.
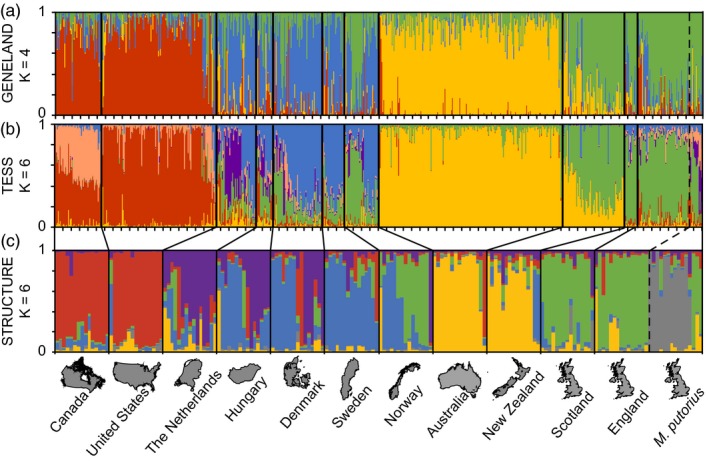
Population assignments (a–c) of ferrets (N = 765) and European polecats (Mustela putorius; N = 15). Program GENELAND (a) identified four genetic clusters whereas programs TESS (b) and STRUCTURE (c) identified six clusters. TESS identified additional substructure in Canada and the Netherlands. On a subsetted dataset with equal sample sizes for each country (N = 15), STRUCTURE assigned most of the European polecats to their own cluster. Polecats were sampled from the United Kingdom and are presented on the far right
The DAPC showed similar patterns to the previous analyses and supports the genetic distinction between ferrets and polecats in the UK (Figure 2). Australia had the least amount of overlap with any other country, indicated by DAPC axis 1 (26.3% of total variation), which primarily separated Australia from all other countries. Polecats clustered most closely to ferrets in the UK, with some overlap. Australian and North American ferrets are most distinct from sampled European polecats, and then ferrets from the UK, indicated on DAPC axis 2 (15.7%). Similar to Bayesian analyses, New Zealand clustered between UK and Australian ferrets, but most closely to the UK. Ferrets from Hungary and Norway also clustered closely to the UK. Ferrets from Denmark, Sweden, and the Netherlands clustered together between UK and North American ferrets.
Figure 2.
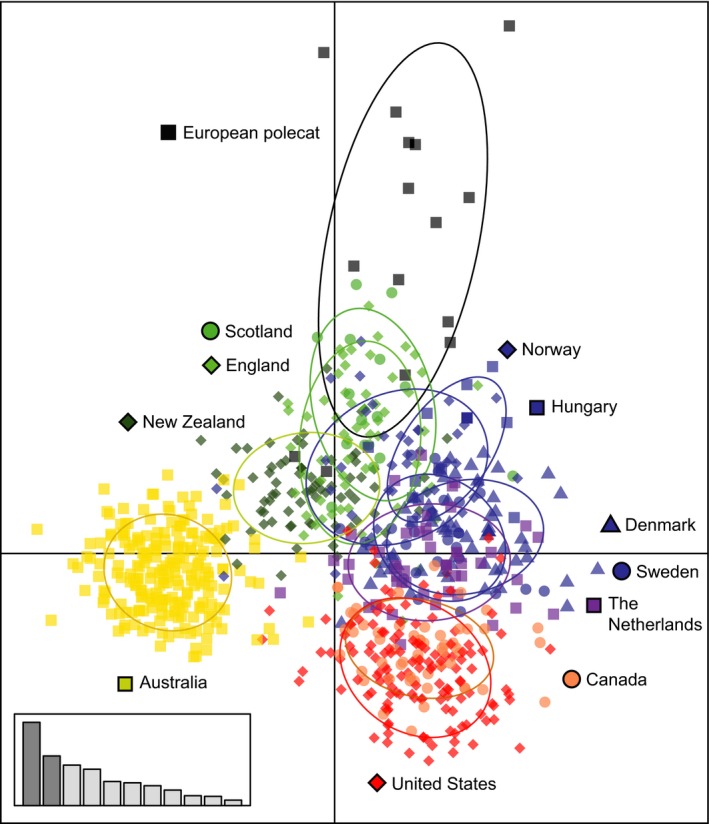
Genetic clustering of ferrets based on a discriminant analysis of principal components. Each dot is an individual ferret or polecat. Each color represents a population identified by program TESS (Figure 1b). European polecats are represented as black squares for easier visualization and based on STRUCTURE analyses (Figure 1c). Discriminant function 1 (x‐axis) accounted for 26.3% of the variation and discriminant function 2 (y‐axis) accounted for 15.7%. The inset barplot shows which axes are being displayed and the relative proportion of variation explained by each of the nine discriminant functions. Two‐thirds of the individuals in each country are contained within the corresponding ellipsoid
3.2. Microsatellite loci and genetic diversity
Based on genetic diversity estimates that are robust to sample size or account for sample size, ferrets from Australia, Canada, and the United States had the lowest genetic diversity (Table 2). Australian ferrets, despite having the largest sample size, had only a single private allele and the lowest number of polymorphic loci. North American ferrets had no private alleles and, with Australia, shared among the lowest measures of allelic richness, Shannon index of allelic diversity, and heterozygosities. Ferrets from the United States and Australia, respectively, had the highest measures of internal relatedness (i.e., inbreeding). New Zealand, England, and Scotland ferrets had intermediate levels of genetic diversity. Ferrets from all other European countries had relatively high estimates of genetic diversity and Hungarian ferrets had the highest allelic richness, number of private alleles, Shannon index, heterozygosities, and the lowest internal relatedness. Notably, ferrets from all countries had mean positive values for internal relatedness, indicative of universal inbreeding.
Table 2.
Genetic diversity of ferrets and European polecats
| Country | N | Ar | SE | PrA | PoL | I | SE | HO | SE | uHE | SE | IR | SE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| European polecat (Mustela putorius) | |||||||||||||
| England | 15 | 3.19 | 0.24 | 2 | 100 | 0.82 | 0.06 | 0.41 | 0.03 | 0.48 | 0.04 | 0.17 | 0.07 |
| Domestic ferret (M. putorius furo) | |||||||||||||
| Australia | 222 | 2.25 | 0.05 | 1 | 90 | 0.54 | 0.05 | 0.30 | 0.03 | 0.32 | 0.03 | 0.23 | 0.01 |
| Canada | 56 | 2.27 | 0.10 | 0 | 94 | 0.56 | 0.05 | 0.34 | 0.03 | 0.35 | 0.03 | 0.16 | 0.02 |
| Denmark | 60 | 3.15 | 0.11 | 8 | 100 | 0.81 | 0.05 | 0.39 | 0.03 | 0.47 | 0.02 | 0.15 | 0.03 |
| England | 63 | 2.90 | 0.10 | 5 | 97 | 0.69 | 0.05 | 0.34 | 0.03 | 0.39 | 0.03 | 0.20 | 0.03 |
| Hungary | 19 | 3.43 | 0.27 | 9 | 97 | 0.83 | 0.06 | 0.43 | 0.03 | 0.48 | 0.03 | 0.07 | 0.03 |
| The Netherlands | 48 | 2.97 | 0.12 | 4 | 100 | 0.78 | 0.05 | 0.38 | 0.03 | 0.46 | 0.03 | 0.16 | 0.03 |
| Norway | 41 | 3.11 | 0.13 | 1 | 100 | 0.78 | 0.05 | 0.40 | 0.02 | 0.45 | 0.03 | 0.11 | 0.04 |
| New Zealand | 74 | 2.63 | 0.09 | 3 | 100 | 0.64 | 0.05 | 0.33 | 0.03 | 0.37 | 0.03 | 0.21 | 0.02 |
| Scotland | 16 | 2.87 | 0.23 | 1 | 97 | 0.67 | 0.05 | 0.39 | 0.04 | 0.41 | 0.03 | 0.10 | 0.05 |
| Sweden | 27 | 2.83 | 0.15 | 2 | 100 | 0.74 | 0.05 | 0.41 | 0.04 | 0.45 | 0.03 | 0.10 | 0.04 |
| United States | 139 | 2.40 | 0.05 | 0 | 100 | 0.56 | 0.05 | 0.30 | 0.03 | 0.33 | 0.03 | 0.25 | 0.02 |
N, sample size; Ar, sample size‐corrected allelic richness; PrA, raw private alleles; PoL, percent of polymorphic loci standardized to sample size; I, Shannon index; HO, observed heterozygosity; uHE, unbiased expected heterozygosity which is robust to sample size differences; IR, average individual internal relatedness based on the entire sample of ferrets and polecats; SE, standard error.
4. DISCUSSION
Intercontinental ferret (Mustela putorius furo) trade has resulted in geographically distinct genetic clusters, likely resulting from founder events and geographic isolation combined with inbreeding and genetic drift. Spatially explicit programs TESS and GENELAND showed comparable population structure, however, TESS identified additional substructure within North America and Hungary. Additionally, our nonspatial STRUCTURE results were highly consistent with those from TESS and GENELAND except that STRUCTURE identified the sampled polecats to be genetically distinct from domestic ferrets, indicating the algorithms from the spatial programs or unequal sample sizes may have overridden the genetic differences between polecats and ferrets in the UK. Within our sample, ferrets within any single country tended to assign to only one of the four genetic clusters, including the United Kingdom (UK), non‐UK Europe, North America, or the Australia cluster. Exceptions to this pattern were New Zealand and Norway. Ferrets in New Zealand primarily assigned to the UK cluster but also shared assignments with the Australian cluster, supporting the hypothesis that New Zealand ferrets originated from England and from Australia (Buller, 1877). Ferrets from Norway either assigned to the UK or the non‐UK European cluster. The four major clusters showed extreme variation in genetic diversity and inbreeding measures. Ferrets in Europe had higher levels of genetic diversity than ferrets on other continents. Australia, Canada, and the United States had ferrets with the lowest genetic diversity and highest inbreeding measures. New Zealand, because of its shared ancestry, had low levels of genetic diversity, but greater diversity than Australian ferrets.
European polecats all exhibited variation at the microsatellite loci developed from the domestic ferret (Ernest et al., 2012), implying common ancestry (Blandford, 1987; Hosoda et al., 2000; Kurose et al., 2000, 2008). However, we did not sample other polecat species and therefore cannot determine whether the domestic ferret was domesticated from other possible species. Identifying the source location for each domestic cluster remains difficult because we do not have samples from each European country where ferrets are present and ferrets in Europe could potentially be hybridized with wild polecats or potentially other species (Costa et al., 2013; Lodé et al., 2005; Pitt, 1921; Poole, 1972), reducing any signal of relationships (Davison et al., 1999; Marmi et al., 2004). Although spatial population assignment models did not differentiate polecats from ferrets, a subsampled dataset with equal sample sizes and uniform priors indicated a genetic distinction, which is consistent with the DAPC.
A previous study observed high expected heterozygosities (mean ± SE: 0.58 ± 0.12) and allelic richness (3.93 ± 0.13) among populations of European polecats from mainland Europe (Pertoldi et al., 2006). Cross‐breeding between ferrets and genetically diverse polecats could explain the high genetic diversity observed in non‐UK European countries. In contrast, polecats in the UK experienced a dramatic reduction in population size during the 19th century (Langley & Yalden, 1977), leading to a genetic bottleneck (Costa et al., 2013). Our observations of genetic diversity for polecats from UK are consistent with previous reports (Costa et al., 2013), which are higher than those of domestic ferrets but lower than polecats from mainland Europe (Moller et al., 2004; Pertoldi et al., 2006). Ferret hybridization is common in the UK but does not seem to be increasing genetic diversity in pet ferrets, possibly because ferret hybridization is occurring with a genetically recovering polecat population that recently went through a bottleneck (Costa et al., 2013). Additionally, three of the 15 polecats assigned as UK ferrets using STRUCTURE, which could indicate we sampled 12 wild polecats and three feral ferrets. Combined, this could explain the high internal relatedness observed in polecats, despite the high genetic diversity. Notably, Hungarian then Danish ferrets had the most private alleles and the highest measures of genetic diversity. Hungary is the only country we sampled with both European polecats (M. putorius) and Steppe polecats (M. eversmanii Lesson, 1872) (Lanszki & Heltai, 2007; Šálek et al., 2013). Similarly, Denmark has both European polecats and American mink (Neovision vison Schreber, 1777) (Hammershøj et al.,2006). Although we do not have direct evidence for hybridization, the mechanism leading to unique alleles in these countries could be a result of hybridization or large effective population sizes.
None of the countries in which we sampled ferrets had an expected heterozygosity (mean among all domestic ferrets in our dataset: 0.41 ± 0.10) or allelic richness (mean among all domestic ferrets in our dataset: 2.80 ± 0.10) approaching those of their wild counterparts (Moller et al., 2004; Pertoldi et al., 2006), indicating founder events, genetic drift, and/or inbreeding have affected ferret genetic diversity (Diamond, 2002; Petersson et al., 1996; Teletchea & Fontaine, 2014). In natural systems, internal relatedness values range from negative (i.e., genetically outbred) to positive (i.e., genetically inbred; Amos et al., 2001). Ferrets from all countries exhibited mean positive internal relatedness, indicative of widespread inbreeding. Artificial selection is well known to reduce genetic diversity (Driscoll et al., 2009; Leroy, 2011; Muñoz‐Fuentes et al., 2014) and breeding for ferret coat color has been shown to be associated with genetically determined physical abnormalities (Blaszczyk et al., 2007; Piazza et al., 2014), which could be an indication of inbreeding depression (Charlesworth & Charlesworth, 1999; Hedrick & Garcia‐Dorado, 2016; Leroy, 2014). Cancer rates are increasing in pet ferret populations (Antinoff & Williams, 2012; Bakthavatchalu, Muthupalani, Marini, & Fox, 2016), and although the mechanism for increasing cancer rates is currently unknown (Fox, Muthupalani, Kiupel, & Williams, 2014), inbreeding and low genetic diversity are suspected (Bakthavatchalu et al., 2016; Bielinska et al., 2009). If lack of genetic diversity is a contributor to cancer acquisition, as in other systems (Epstein et al., 2016; McAloose & Newton, 2009; Morris, Wright, Grueber, Hogg, & Belov, 2015; Rahman, 2014), ferrets in Australia, Canada, New Zealand, and the United States are most at risk.
One of the primary ways to reverse inbreeding depression is through genetic restoration (Frankham, 2015; Whiteley, Fitzpatrick, Funk, & Tallmon, 2015). In domestic animal populations, genetic restoration requires human intervention and breeding programs (Ralls & Ballou, 1986; Rollinson et al., 2014). However, international trade laws could limit the feasibility of introducing new genetic material to genetically depauperate pet ferret populations, especially on continents without potential for interbreeding with wild polecats. For example, ferrets are common pets in Australia (Talbot, Freire, & Wassens, 2014) and our study supports the assertion that inbreeding has been a national problem (Lewington, 2007b). However, ferret importation is currently illegal in Australia (Department of the Environment and Energy, 2017) and ferrets are completely prohibited in Queensland (Queensland Government, 2016) and the Northern Territory (Northern Territory Government, 2016). Despite New Zealand allowing ferret exportation for permitted breeders, the purchase and importation of ferrets were banned because of damage to native species (Lee, 2002; O'Donnell et al., 2017; Wells, 2009). Thus, Australia and New Zealand breeders should actively minimize inbreeding among currently available ferrets.
We do not see any particular utility for maintaining genetically distinct ferret clusters in most cases. Thus, for countries with inbred ferrets, we recommend that pet and laboratory breeding programs should incorporate ferrets from other countries. Although outbreeding depression could result from such crosses, the potential for inbreeding depression is much more likely (Rollinson et al., 2014). Additionally, it could also be genetically beneficial for breeding programs to introduce new genes into their domestic lines through ethical and legal cross‐breeding with wild polecats. However, this could potentially result in ferrets with different behaviors than those that are considered desirable for pets (Hernádi et al., 2012) or the introduction of unwanted diseases. If this action is taken, breeders should ensure ferrets do not breed into the wild population, which could put the wild population at risk of genetic disorders (Costa et al., 2013; Rhymer & Simberloff, 1996). To reduce potential risk of inbreeding depression, ferret breeding programs designed for specific coat colors should consider mating unrelated individuals and occasionally “diluting” specific coat‐color lines with individuals of different varieties.
The United States and Canada allow ferret importation (Animal and Plant Health Inspection Service, 2016; Canadian Food Inspection Agency, 2015), yet have among the lowest measures of genetic diversity observed. Despite the large sample size, North American ferrets had no private alleles, providing no evidence of hybridization with native black‐footed ferrets (Williams et al., 1996). The opportunity for increasing domestic ferret genetic diversity in North America is currently available and should be considered given the application of ferrets as model organisms in North American biomedical laboratories (Ball, 2006; Jones et al., 2017; Peng et al., 2014; Porter, 2016; Vanchieri, 2001). Researchers should consider the effect low genetic diversity and inbreeding can have on the results and inferences from disease exposure experiments (Spielman, Brook, Briscoe, & Frankham, 2004; Whiteman, Matson, Bollmer, & Parker, 2006). Although causal links between neutral genetic markers and neoplasms cannot be made, our research highlights the lack of genetic variation present in certain ferret clusters. With the sequencing of the ferret genome being recently completed (Peng et al., 2014), researchers can now search the genome for specific genes which may be linked to cancer acquisition in this developing cancer model system (Aizawa et al., 2013, 2016).
Overall, our study identified international and intercontinental domestic ferret population structure and indicates Australian and North American ferrets have the lowest genetic diversities and are most highly diverged from the European polecat. Given current importation bans, New Zealand ferrets are essentially isolated and, in the future, may also exhibit patterns of low genetic diversity and inbreeding. Prevention or mitigation of inbreeding depression will require international cooperation among breeding programs and should include genetically diverse ferrets from mainland Europe. In some cases, political factors limit the ability of potential international breeding programs. This research highlights the need for politicians, coat‐oriented breeders, and laboratory‐stock breeders to reassess their policies. Given the close relatedness among domestic ferrets, European polecats, Steppe polecats, black‐footed ferrets, American mink, and European mink (Cabria et al., 2011; Kurose et al., 2008; Lodé et al., 2005; Williams et al., 1996), the conservation genetics approach used in this study has practical applications for conserving the genetic diversity of related wild species that could face reduction in genetic diversity when hybridized with feral ferrets (Bonesi & Palazon, 2007; Cabria et al., 2015; Šálek et al., 2013; Wisely, Buskirk, Fleming, McDonald, & Ostrander, 2002) and for the identification and management of feral ferret source populations causing ecosystem damage (Bodey, Bearhop, & McDonald, 2011; Byrom, Caley, Paterson, & Nugent, 2015; O'Donnell et al., 2017; Wells, 2009).
DATA ARCHIVING STATEMENT
Genotype data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.24c24
Supporting information
ACKNOWLEDGEMENTS
We thank N. Pederson for providing early ideas. This project was supported by the Center for Companion Animal Health, School of Veterinary Medicine, University of California, Davis, USA (07‐04‐F).
Gustafson KD, Hawkins MG, Drazenovich TL, Church R, Brown SA, Ernest HB. Founder events, isolation, and inbreeding: Intercontinental genetic structure of the domestic ferret. Evol Appl. 2018;11:694–704. https://doi.org/10.1111/eva.12565
REFERENCES
- Aizawa, K. , Liu, C. , Tang, S. , Veeramachaneni, S. , Hu, K. Q. , Smith, D. E. , & Wang, X. D. (2016). Tobacco carcinogen induces both lung cancer and non‐alcoholic steatohepatitis and hepatocellular carcinomas in ferrets which can be attenuated by lycopene supplementation. International Journal of Cancer, 139, 1171–1181. https://doi.org/10.1002/ijc.30161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa, K. , Liu, C. , Veeramachaneni, S. , Hu, K.‐Q. , Smith, D. E. , & Wang, X.‐D. (2013). Development of ferret as a human lung cancer model by injecting 4‐(N‐methyl‐N‐nitrosamino)‐1‐(3‐pyridyl)‐1‐butanone (NNK). Lung Cancer, 82, 390–396. https://doi.org/10.1016/j.lungcan.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alho, J. S. , Valimaki, K. , & Merila, J. (2010). Rhh: An R extension for estimating multilocus heterozygosity and heterozygosity‐heterozygosity correlation. Molecular Ecology Resources, 10, 720–722. https://doi.org/10.1111/j.1755-0998.2010.02830.x [DOI] [PubMed] [Google Scholar]
- Amos, W. , Wilmer, J. W. , Fullard, K. , Burg, T. , Croxall, J. , Bloch, D. , & Coulson, T. (2001). The influence of parental relatedness on reproductive success. Proceedings of the Royal Society of London B: Biological Sciences, 268, 2021–2027. https://doi.org/10.1098/rspb.2001.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Animal and Plant Health Inspection Service (2016). Pet Travel – Bringing Ferrets into the US. United States Department of Agriculture Retrieved from https://www.aphis.usda.gov/aphis/pet-travel/bring-pet-into-the-united-states/pet-travel-ferrets-into-us (Archived by WebCite® at http://www.webcitation.org/6oHRaEFtJ)
- Antinoff, N. , & Williams, B. (2012). Neoplasia In Quesenberry K. E. & Carpenter J. L. (Eds.), Ferrets, rabbits and rodents: Clinical medicine and surgery (3rd ed., pp. 103–122). St Louis, MO: Elsevier; https://doi.org/10.1016/B978-1-4160-6621-7.00008-7 [Google Scholar]
- Avallone, G. , Forlani, A. , Tecilla, M. , Riccardi, E. , Belluco, S. , Santagostino, S. F. , … Roccabianca, P. (2016). Neoplastic diseases in the domestic ferret (Mustela putorius furo) in Italy: Classification and tissue distribution of 856 cases (2000–2010). BMC Veterinary Research, 12, 275 https://doi.org/10.1186/s12917-016-0901-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatchalu, V. , Muthupalani, S. , Marini, R. , & Fox, J. (2016). Endocrinopathy and aging in ferrets. Veterinary Pathology, 53, 349–365. https://doi.org/10.1177/0300985815623621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, R. S. (2006). Issues to consider for preparing ferrets as research subjects in the laboratory. ILAR Journal, 47, 348–357. https://doi.org/10.1093/ilar.47.4.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinska, M. , Parviainen, H. , Kiiveri, S. , Heikinheimo, M. , & Wilson, D. B. (2009). Review paper: Origin and molecular pathology of adrenocortical neoplasms. Veterinary Pathology, 46, 194–210. https://doi.org/10.1354/vp.46-2-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandford, P. (1987). Biology of the polecat Mustela putorius: A literature review. Mammal Review, 17, 155–198. https://doi.org/10.1111/j.1365-2907.1987.tb00282.x [Google Scholar]
- Blaszczyk, W. M. , Distler, C. , Dekomien, G. , Arning, L. , Hoffmann, K. P. , & Epplen, J. T. (2007). Identification of a tyrosinase (TYR) exon 4 deletion in albino ferrets (Mustela putorius furo). Animal Genetics, 38, 421–423. https://doi.org/10.1111/j.1365-2052.2007.01619.x [DOI] [PubMed] [Google Scholar]
- Bodey, T. W. , Bearhop, S. , & McDonald, R. A. (2011). The diet of an invasive nonnative predator, the feral ferret Mustela furo, and implications for the conservation of ground‐nesting birds. European Journal of Wildlife Research, 57, 107–117. https://doi.org/10.1007/s10344-010-0404-y [Google Scholar]
- Bonesi, L. , & Palazon, S. (2007). The American mink in Europe: Status, impacts, and control. Biological Conservation, 134, 470–483. https://doi.org/10.1016/j.biocon.2006.09.006 [Google Scholar]
- Buller, C. M. G. (1877). On the proposed introduction of the polecat into New Zealand. Transactions and Proceedings of the Royal Society of New Zealand, 9, 634–635. http://rsnz.natlib.govt.nz/volume/rsnz_09.html [Google Scholar]
- Byrom, A. E. , Caley, P. , Paterson, B. M. , & Nugent, G. (2015). Feral ferrets (Mustela furo) as hosts and sentinels of tuberculosis in New Zealand. New Zealand Veterinary Journal, 63, 42–53. https://doi.org/10.1080/00480169.2014.981314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabria, M. T. , Gonzalez, E. G. , Gomez‐Moliner, B. J. , Michaux, J. R. , Skumatov, D. , Kranz, A. , … Zardoya, R. (2015). Patterns of genetic variation in the endangered European mink (Mustela lutreola L., 1761). BMC Evolutionary Biology, 15, 141 https://doi.org/10.1186/s12862-015-0427-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabria, M. T. , Michaux, J. R. , GóMez‐Moliner, B. J. , Skumatov, D. , Maran, T. , Fournier, P. , … Zardoya, R. (2011). Bayesian analysis of hybridization and introgression between the endangered European mink (Mustela lutreola) and the polecat (Mustela putorius). Molecular Ecology, 20, 1176–1190. https://doi.org/10.1111/j.1365-294x.2010.04988.x [DOI] [PubMed] [Google Scholar]
- Canadian Food Inspection Agency (2015). Importing or travelling with foxes, skunks, raccoons and ferrets as pets. Government of Canada Retrieved from http://www.inspection.gc.ca/animals/terrestrial-animals/imports/policies/live-animals/pets/ferrets-etc-/eng/1331923269633/1331923405610 (Archived by WebCite® at http://www.webcitation.org/6oHRIRG05)
- Carnegie, W. (2013). Ferrets & ferreting‐a practical manual on breeding, managing, training and working ferrets. Redditch, United Kingdom: Read Books Ltd. [Google Scholar]
- Charlesworth, B. , & Charlesworth, D. (1999). The genetic basis of inbreeding depression. Genetical Research, 74, 329–340. https://doi.org/10.1017/S0016672399004152 [DOI] [PubMed] [Google Scholar]
- Church, B. (2007). Ferret‐polecat domestication: Genetic, taxonomic and phylogenetic relationships In Lewington J. H. (Ed.), Ferret husbandry, medicine and surgery (Vol. 2, pp. 122–150). Edinburgh, UK: Saunders Elsevier; https://doi.org/10.1016/B978-0-7020-2827-4.50012-7 [Google Scholar]
- Clagett, D. O. , Johnston, M. S. , & Han, S. (2016). Malignant plasma cell neoplasia in ferrets: A review of six cases. Journal of Exotic Pet Medicine, 26, 36–46. https://doi.org/10.1053/j.jepm.2016.10.010 [Google Scholar]
- Costa, M. , Fernandes, C. , Birks, J. D. S. , Kitchener, A. C. , Santos‐Reis, M. , & Bruford, M. W. (2013). The genetic legacy of the 19th‐century decline of the British polecat: Evidence for extensive introgression from feral ferrets. Molecular Ecology, 22, 5130–5147. https://doi.org/10.1111/mec.12456 [DOI] [PubMed] [Google Scholar]
- Dallas, J. , & Piertney, S. (1998). Microsatellite primers for the Eurasian otter. Molecular Ecology, 7, 1248–1251. [PubMed] [Google Scholar]
- Davison, A. , Birks, J. , Griffiths, H. , Kitchener, A. , Biggins, D. , & Butlin, R. (1999). Hybridization and the phylogenetic relationship between polecats and domestic ferrets in Britain. Biological Conservation, 87, 155–161. https://doi.org/10.1016/S0006-3207(98)00067-6 [Google Scholar]
- Department of the Environment and Energy (2017). Live import list: amendments to the live import list. Australian Government. Retrieved from http://www.environment.gov.au/biodiversity/wildlife-trade/live/import-list/amendments (Archived by WebCite® at http://www.webcitation.org/6oHNwmQ7c)
- Diamond, J. (2002). Evolution, consequences and future of plant and animal domestication. Nature, 418, 700–707. https://doi.org/10.1038/nature01019 [DOI] [PubMed] [Google Scholar]
- Domingo‐Roura, X. , Macdonald, D. , Roy, M. , Marmi, J. , Terradas, J. , Woodroffe, R. , … Wayne, R. (2003). Confirmation of low genetic diversity and multiple breeding females in a social group of Eurasian badgers from microsatellite and field data. Molecular Ecology, 12(2), 533–540. https://doi.org/10.1046/j.1365-294X.2003.01707.x [DOI] [PubMed] [Google Scholar]
- Driscoll, C. A. , Macdonald, D. W. , & O'Brien, S. J. (2009). From wild animals to domestic pets, an evolutionary view of domestication. Proceedings of the National Academy of Sciences, 106, 9971–9978. https://doi.org/10.1073/pnas.0901586106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, E. , Chen, C. , & François, O. (2009). TESS version 2.3 Reference Manual.
- Epstein, B. , Jones, M. , Hamede, R. , Hendricks, S. , McCallum, H. , Murchison, E. P. , … Storfer, A. (2016). Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nature Communications, 7, 12684 https://doi.org/10.1038/ncomms12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernest, H. B. , Drazenovich, T. L. , Dalbeck, L. S. , & Hawkins, M. G. (2012). Isolation and characterization of microsatellite markers in the domestic ferret (Mustela putorius furo). International Journal of Molecular Sciences, 13, 16592–16597. https://doi.org/10.3390/ijms131216592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, J. G. , & Marini, R. P. (2014). Biology and diseases of the ferret (Vol. 2). Ames, IA: John Wiley & Sons; https://doi.org/10.1002/9781118782699 [Google Scholar]
- Fox, J. G. , Muthupalani, S. , Kiupel, M. , & Williams, B. (2014). Neoplastic diseases In Fox J. G. & Marini R. P. (Eds.), Biology and diseases of the ferret (Vol. 2, pp. 587–626). New York, NY: John Wiley & Sons; https://doi.org/10.1002/9781118782699 [Google Scholar]
- Frankham, R. (2015). Genetic rescue of small inbred populations: Meta‐analysis reveals large and consistent benefits of gene flow. Molecular Ecology, 24, 2610–2618. https://doi.org/10.1111/mec.13139 [DOI] [PubMed] [Google Scholar]
- González‐Recio, O. , López de Maturana, E. , & Gutiérrez, J. P. (2007). Inbreeding depression on female fertility and calving ease in Spanish dairy cattle. Journal of Dairy Science, 90, 5744–5752. https://doi.org/10.3168/jds.2007-0203 [DOI] [PubMed] [Google Scholar]
- Goudet, J. (1995). FSTAT (version 1.2): A computer program to calculate F‐statistics. Journal of Heredity, 86, 485–486. https://doi.org/10.1093/oxfordjournals.jhered.a111627 [Google Scholar]
- Guillot, G. , Estoup, A. , Mortier, F. , & Cosson, J. F. (2005). A spatial statistical model for landscape genetics. Genetics, 170, 1261–1280. https://doi.org/10.1534/genetics.104.033803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot, G. , Mortier, F. , & Estoup, A. (2005). GENELAND: A computer package for landscape genetics. Molecular Ecology Notes, 5, 712–715. https://doi.org/10.1111/j.1471-8286.2005.01031.x [Google Scholar]
- Hammershøj, M. , Travis, J. M. J. , & Stephenson, C. M . (2006). Incorporating evolutionary processes into a spatially‐explicit model: Exploring the consequences of mink‐farm closures in Denmark. Ecography, 29, 465–476. https://doi.org/10.1111/j.2006.0906-7590.04492.x [Google Scholar]
- Hedrick, P. W. , & Garcia‐Dorado, A. (2016). Understanding inbreeding depression, purging, and genetic rescue. Trends in Ecology & Evolution, 31, 940–952. https://doi.org/10.1016/j.tree.2016.09.005 [DOI] [PubMed] [Google Scholar]
- Hernádi, A. , Kis, A. , Turcsán, B. , & Topál, J. (2012). Man's underground best friend: Domestic ferrets, unlike the wild forms, show evidence of dog‐like social‐cognitive skills. PLoS ONE, 7, e43267 https://doi.org/10.1371/journal.pone.0043267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda, T. , Suzuki, H. , Harada, M. , Tsuchiya, K. , Han, S.‐H. , Zhang, Y.‐P. , … Lin, L.‐K. (2000). Evolutionary trends of the mitochondrial lineage differentiation in species of genera Martes and Mustela. Genes & Genetic Systems, 75, 259–267. https://doi.org/10.1266/ggs.75.259 [DOI] [PubMed] [Google Scholar]
- Howard, J. , Lynch, C. , Santymire, R. , Marinari, P. , & Wildt, D. (2015). Recovery of gene diversity using long‐term cryopreserved spermatozoa and artificial insemination in the endangered black‐footed ferret. Animal Conservation, 19, 102–111. https://doi.org/10.1111/acv.12229 [Google Scholar]
- Ilmonen, P. , Penn, D. J. , Damjanovich, K. , Clarke, J. , Lamborn, D. , Morrison, L. , … Potts, W. K. (2008). Experimental infection magnifies inbreeding depression in house mice. Journal of Evolutionary Biology, 21, 834–841. https://doi.org/10.1111/j.1420-9101.2008.01510.x [DOI] [PubMed] [Google Scholar]
- Jakobsson, M. , & Rosenberg, N. A. (2007). CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801–1806. https://doi.org/10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- Jombart, T. (2008). adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403–1405. https://doi.org/10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Jombart, T. , Devillard, S. , & Balloux, F. (2010). Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genetics, 11, 94 https://doi.org/10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, B. , Donovan, C. , Liu, G. , Gomez, H. M. , Chimankar, V. , Harrison, C. L. , … Hirota, J. A. (2017). Animal models of COPD: What do they tell us? Respirology, 22, 21–32. https://doi.org/10.1111/resp.12908 [DOI] [PubMed] [Google Scholar]
- Kalinowski, S. T. (2004). Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conservation Genetics, 5, 539–543. https://doi.org/10.1023/B:COGE.0000041021.91777.1a [Google Scholar]
- Kurose, N. , Abramov, A. V. , & Masuda, R. (2000). Intrageneric diversity of the cytochrome b gene and phylogeny of Eurasian species of the genus Mustela (Mustelidae, Carnivora). Zoological Science, 17, 673–679. https://doi.org/10.2108/zsj.17.673 [DOI] [PubMed] [Google Scholar]
- Kurose, N. , Abramov, A. V. , & Masuda, R. (2008). Molecular phylogeny and taxonomy of the genus Mustela (Mustelidae, Carnivora), inferred from mitochondrial DNA sequences: New perspectives on phylogenetic status of the back‐striped weasel and American mink. Mammal Study, 33, 25–33. http://doi.org/10.3106/1348-6160(2008)33%5b25:mpatot%5d2.0.co;2 [Google Scholar]
- Lam, L. , Gagne, R. B. , & Ernest, H. B. (2016). Development of 24 polymorphic microsatellite loci for the threatened Southern (California) Sea otter (Enhydra lutris nereis) In Arias M. C., Aulagnier S., Baerwald E. F., Barclay R. M. R., Batista J. S., Beasley R. R., … Zou S. (Eds.), Microsatellite records for volume 8, issue 1 (Vol. 8, pp. 1–4): Conservation Genetics Resources, 8, 1–4. https://doi.org/10.1007/s12686-016-0522-2 [Google Scholar]
- Langley, P. , & Yalden, D. (1977). The decline of the rarer carnivores in Great Britain during the nineteenth century. Mammal Review, 7, 95–116. https://doi.org/10.1111/j.1365-2907.1977.tb00363.x [Google Scholar]
- Lanszki, J. , & Heltai, M. (2007). Diet of the European polecat and the steppe polecat in Hungary. Mammalian Biology, 72, 49–53. https://doi.org/10.1016/j.mambio.2006.07.002 [Google Scholar]
- Larson, G. , & Burger, J. (2013). A population genetics view of animal domestication. Trends in Genetics, 29, 197–205. https://doi.org/10.1016/j.tig.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Lee, S. (2002). Pet ferrets to be banned. The Official Website of the New Zealand Government Retrieved from https://www.beehive.govt.nz/release/pet-ferrets-be-banned (Archived by WebCite® at http://www.webcitation.org/6oB0BUqUk)
- Leroy, G. (2011). Genetic diversity, inbreeding and breeding practices in dogs: Results from pedigree analyses. The Veterinary Journal, 189, 177–182. https://doi.org/10.1016/j.tvjl.2011.06.016 [DOI] [PubMed] [Google Scholar]
- Leroy, G. (2014). Inbreeding depression in livestock species: Review and meta‐analysis. Animal Genetics, 45, 618–628. https://doi.org/10.1111/age.12178 [DOI] [PubMed] [Google Scholar]
- Lewington, J. H. (2007a). Classification, history and current status of ferrets In Lewington J. (Ed.), Ferret husbandry, medicine and surgery (Vol. 2, pp. 3–14). Edinburgh, UK: Saunders Elsevier. [Google Scholar]
- Lewington, J. H. (2007b). Reproduction and genetics In Lewington J. (Ed.), Ferret husbandry, medicine and surgery (Vol. 2, pp. 86–121). Edinburgh, UK: Saunders Elsevier. [Google Scholar]
- Lodé, T. , Guiral, G. , & Peltier, D. (2005). European mink–polecat hybridization events: Hazards from natural process? Journal of Heredity, 96, 89–96. https://doi.org/10.1093/jhered/esi021 [DOI] [PubMed] [Google Scholar]
- Marmi, J. , López‐Giráldez, J. F. , & Domingo‐Roura, X. (2004). Phylogeny, evolutionary history and taxonomy of the Mustelidae based on sequences of the cytochrome b gene and a complex repetitive flanking region. Zoologica Scripta, 33, 481–499. https://doi.org/10.1111/j.0300-3256.2004.00165.x [Google Scholar]
- McAloose, D. , & Newton, A. L. (2009). Wildlife cancer: A conservation perspective. Nature Reviews Cancer, 9, 517–526. https://doi.org/10.1038/nrc2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, M. , Yahara, T. , Faith, D. P. , Vázquez‐Domínguez, E. , Colautti, R. I. , Araki, H. , … Hendry, A. P. (2017). Understanding and monitoring the consequences of human impacts on intraspecific variation. Evolutionary Applications, 10, 121–139. https://doi.org/10.1111/eva.12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, T. , Pertoldi, C. , Madsen, A. B. , Asferg, T. , Frydenberg, J. , Hammershoj, M. , & Loeschcke, V. (2004). Genetic variability in Danish polecats Mustela putorius as assessed by microsatellites. Wildlife Biology, 10, 25–34. [Google Scholar]
- Morris, K. M. , Wright, B. , Grueber, C. E. , Hogg, C. , & Belov, K. (2015). Lack of genetic diversity across diverse immune genes in an endangered mammal, the Tasmanian devil (Sarcophilus harrisii). Molecular Ecology, 24, 3860–3872. https://doi.org/10.1111/mec.13291 [DOI] [PubMed] [Google Scholar]
- Muñoz‐Fuentes, V. , Marcet‐Ortega, M. , Alkorta‐Aranburu, G. , Linde Forsberg, C. , Morrell, J. M. , Manzano‐Piedras, E. , … Vilà, C. (2014). Strong artificial selection in domestic mammals did not result in an increased recombination rate. Molecular Biology and Evolution, 32, 510–523. https://doi.org/10.1093/molbev/msu322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northern Territory Government (2016). Prohibited wildlife. Northern Territory Government of Australia Retrieved from https://nt.gov.au/environment/animals/wildlife-permits/prohibited-wildlife (Archived by WebCite® at http://www.webcitation.org/6oB9oKaMM)
- O'Connell, M. , Wright, J. M. , & Farid, A. (1996). Development of PCR primers for nine polymorphic American mink Mustela vison microsatellite loci. Molecular Ecology, 5, 311–312. https://doi.org/10.1046/j.1365-294X.1996.00103.x [DOI] [PubMed] [Google Scholar]
- O'Donnell, C. F. , Weston, K. A. , & Monks, J. M. (2017). Impacts of introduced mammalian predators on New Zealand's alpine fauna. New Zealand Journal of Ecology, 41(1), 01–22. https://doi.org/10.20417/nzjecol [Google Scholar]
- O'Neill, D. , Church, D. , McGreevy, P. , Thomson, P. , & Brodbelt, D. (2013). Longevity and mortality of owned dogs in England. The Veterinary Journal, 198, 638–643. https://doi.org/10.1016/j.tvjl.2013.09.020 [DOI] [PubMed] [Google Scholar]
- Owen, C. (1984). Ferret In Mason I. L. (Ed.), Evolution of domesticated animals (pp. 225–228). London, UK: Longman. [Google Scholar]
- Paetkau, D. , & Strobeck, C. (1994). Microsatellite analysis of genetic variation in black bear populations. Molecular Ecology, 3(5), 489–495. https://doi.org/10.1111/j.1365-294X.1994.tb00127.x [DOI] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2006). GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics, 28, 2537–2539. https://doi.org/10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, X. , Alfoldi, J. , Gori, K. , Eisfeld, A. J. , Tyler, S. R. , Tisoncik‐Go, J. , … Katze, M. G. (2014). The draft genome sequence of the ferret (Mustela putorius furo) facilitates study of human respiratory disease. Nature Biotechnology, 32, 1250–1255. https://doi.org/10.1038/nbt.3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertoldi, C. , Breyne, P. , Cabria, M. T. , Halfmaerten, D. , Jansman, H. A. H. , Van Den Berge, K. , … Loeschcke, V. (2006). Genetic structure of the European polecat (Mustela putorius) and its implication for conservation strategies. Journal of Zoology, 270, 102–115. https://doi.org/10.1111/j.1469-7998.2006.00095.x [Google Scholar]
- Petersson, E. , Jaurvi, T. , Steffner, N. G. , & Ragnarsson, B. (1996). The effect of domestication on some life history traits of sea trout and Atlantic salmon. Journal of Fish Biology, 48, 776–791. https://doi.org/10.1111/j.1095-8649.1996.tb01471.x [Google Scholar]
- Piazza, S. , Abitbol, M. , Gnirs, K. , Huynh, M. , & Cauzinille, L. (2014). Prevalence of deafness and association with coat variations in client‐owned ferrets. Journal of the American Veterinary Medical Association, 244, 1047–1052. https://doi.org/10.2460/javma.244.9.1047 [DOI] [PubMed] [Google Scholar]
- Pitt, F. (1921). Notes on the genetic behaviour of certain characters in the polecat, ferret, and in polecat‐ferret hybrids. Journal of Genetics, 11, 95–115. https://doi.org/10.1007/BF02983044 [Google Scholar]
- Poole, T. B. (1972). Some behavioural differences between the European polecat, Mustela putorius, the ferret, M. furo, and their hybrids. Journal of Zoology, 166, 25–35. [Google Scholar]
- Porter, N. H. (2016). Ferreting things out: Biosecurity, pandemic flu and the transformation of experimental systems. BioSocieties, 11, 22–45. https://doi.org/10.1057/biosoc.2015.4 [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P . (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puechmaille, S. J. (2016). The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Molecular Ecology Resources, 16, 608–627. https://doi.org/10.1111/1755-0998.12512 [DOI] [PubMed] [Google Scholar]
- Queensland Government (2016). Prohibited invasive animals: Ferret. The State of Queensland 1995–2017 Retrieved from https://www.business.qld.gov.au/industries/farms-fishing-forestry/agriculture/land-management/health-pests-weeds-diseases/pests/invasive-animals/prohibited/ferret (Archived by WebCite® at http://www.webcitation.org/6oB6x1FVk)
- Rahman, N. (2014). Realizing the promise of cancer predisposition genes. Nature, 505, 302–308. https://doi.org/10.1038/nature12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralls, K. , & Ballou, J. (1986). Captive breeding programs for populations with a small number of founders. Trends in Ecology & Evolution, 1, 19–22. https://doi.org/10.1016/0169-5347(86)90062-5 [DOI] [PubMed] [Google Scholar]
- Rhymer, J. M. , & Simberloff, D. (1996). Extinction by hybridization and introgression. Annual Review of Ecology and Systematics, 27, 83–109. https://doi.org/10.1146/annurev.ecolsys.27.1.83 [Google Scholar]
- Rollinson, N. , Keith, D. M. , Houde, A. L. S. , Debes, P. V. , McBride, M. C. , & Hutchings, J. A. (2014). Risk assessment of inbreeding and outbreeding depression in a captive‐breeding program. Conservation Biology, 28, 529–540. https://doi.org/10.1111/cobi.12188 [DOI] [PubMed] [Google Scholar]
- Rousset, F. (2008). GENEPOP'007: A complete re‐implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources, 8, 103–106. https://doi.org/10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- Šálek, M. , Spassov, N. , Anděra, M. , Enzinger, K. , Ottlecz, B. , & Hegyeli, Z. (2013). Population status, habitat associations, and distribution of the steppe polecat Mustela eversmanii in Europe. Acta Theriologica, 58, 233–244. https://doi.org/10.1007/s13364-013-0134-0 [Google Scholar]
- Spielman, D. , Brook, B. W. , Briscoe, D. A. , & Frankham, R. (2004). Does inbreeding and loss of genetic diversity decrease disease resistance? Conservation Genetics, 5, 439–448. https://doi.org/10.1023/B:COGE.0000041030.76598.cd [Google Scholar]
- Talbot, S. , Freire, R. , & Wassens, S. (2014). Effect of captivity and management on behaviour of the domestic ferret (Mustela putorius furo). Applied Animal Behaviour Science, 151, 94–101. https://doi.org/10.1016/j.applanim.2013.11.017 [Google Scholar]
- Teletchea, F. , & Fontaine, P. (2014). Levels of domestication in fish: Implications for the sustainable future of aquaculture. Fish and Fisheries, 15, 181–195. https://doi.org/10.1111/faf.12006 [Google Scholar]
- Thomson, A. P. D. (1951). A history of the ferret. Journal of the History of Medicine and Allied Sciences, 6, 471–480. https://doi.org/10.1093/jhmas/VI.Autumn.471 [Google Scholar]
- Toonen, R. J. , & Hughes, S. (2001). Increased throughput for fragment analysis on an ABI Prism® 377 automated sequencer using a membrane comb and STRand software. BioTechniques, 31, 1320–1325. [PubMed] [Google Scholar]
- Vanchieri, C. (2001). Move over, mouse: Make way for the woodchucks, ferrets, and zebrafish. JNCI: Journal of the National Cancer Institute, 93, 418–419. https://doi.org/10.1093/jnci/93.6.418 [DOI] [PubMed] [Google Scholar]
- Wells, P. (2009). The fall and fall in the legal status of mustelids in New Zealand. Environment and History, 15, 343–368. https://doi.org/10.3197/096734009X12474738225593 [Google Scholar]
- Whiteley, A. R. , Fitzpatrick, S. W. , Funk, W. C. , & Tallmon, D. A. (2015). Genetic rescue to the rescue. Trends in Ecology & Evolution, 30, 42–49. https://doi.org/10.1016/j.tree.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Whiteman, N. K. , Matson, K. D. , Bollmer, J. L. , & Parker, P. G. (2006). Disease ecology in the Galápagos Hawk (Buteo galapagoensis): Host genetic diversity, parasite load and natural antibodies. Proceedings of the Royal Society B: Biological Sciences, 273, 797–804. https://doi.org/10.1098/rspb.2005.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, E. , Anderson, S. , Cavender, J. , Lynn, C. , List, K. , Hearn, C. , & Appel, M. (1996). Vaccination of black‐footed ferret (Mustela nigripes) × Siberian polecat (M. eversmanni) hybrids and domestic ferrets (M. putorius furo) against canine distemper. Journal of Wildlife Diseases, 32, 417–423. https://doi.org/10.7589/0090-3558-32.3.417 [DOI] [PubMed] [Google Scholar]
- Willoughby, J. R. , Fernandez, N. B. , Lamb, M. C. , Ivy, J. A. , Lacy, R. C. , & DeWoody, J. A. (2015). The impacts of inbreeding, drift and selection on genetic diversity in captive breeding populations. Molecular Ecology, 24, 98–110. https://doi.org/10.1111/mec.13020 [DOI] [PubMed] [Google Scholar]
- Wisely, S. M. , Buskirk, S. W. , Fleming, M. A. , McDonald, D. B. , & Ostrander, E. A. (2002). Genetic diversity and fitness in black‐footed ferrets before and during a bottleneck. Journal of Heredity, 93, 231–237. https://doi.org/10.1093/jhered/93.4.231 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


