Abstract
Dogs are competent reservoir hosts of several zoonotic agents, including Filariidae nematodes and Anaplasmataceae family bacteria. The latter family unites human and veterinary pathogens (Anaplasma, Ehrlichia and Neorickettsia bacteria) with Wolbachia, some of which are obligatory endosymbionts of pathogenic filarial nematodes. The epidemiology of Anaplasmataceae and Filariidae species infecting dogs living in kennels in New Caledonia was studied. 64 EDTA blood samples were screened for the presence of Anaplasmataceae and filarial nematodes. Molecular study was conducted using primers and probe targeting the of 23S rRNA long fragment of Anaplasmataceae species. Next, all blood sample was screened for the presence of Filariidae species targeting the primers and probe targeting the COI gene, as well as primers targeting the COI and 5S rRNA genes of all filarial worms. Anaplasma platys was identified in 8/64 (12.5, 95% confidence interval [CI]: 4.4–20.6%) and Wolbachia endosymbiont of Dirofilaria immitis in 8/64 (12.5%, CI: 4.4–20.6%). Filariidae species investigation was performed and showed that 11/64 (17.2%, CI: 7.9–26.4%) dogs were infected with D. immitis, whereas, 2/64 (3.1%, CI: 0.0–7.3%) were infected with Acanthocheilonema reconditum. Finally, we checked the occurrence of co‐infection between Anaplasmataceae and Filariidae species. Co‐occurrence with Wolbachia endosymbiont of D. immitis was observed in seven dogs, one dog was co‐infected with A. platys and A. reconditum and another was co‐infected with Wolbachia endosymbiont of D. immitis and A. reconditum. These results are the first report of Anaplasmataceae and Filariidae occurring in dogs in New Caledonia.
Keywords: Dogs, kennels, New Caledonia, Anaplasmataceae, Filariidae
Introduction
The Anaplasmataceae family of bacteria includes several dog‐associated pathogens such as Anaplasma phagocytophilum, A. platys and Ehrlichia canis, E. chaffeensis, E. ewingii and Neorickettsia helminthoeca (Dumler et al. 2001). Canine granulocytic anaplasmosis due to A. phagocytophilum is reported worldwide and transmitted by Ixodes ricinus complex ticks in Europe and North America (Dumler et al. 2005; Kohn et al. 2011). Anaplasma phagocytophilum and E. canis are the causative agents of cyclic thrombocytopenia and canine monocyte tropic ehrlichiosis, and are reported worldwide. Infections are often found in geographical regions associated with the Rhipicephalus sanguineus s.l. tick (Otranto et al. 2009; Harrus & Waner 2011). Ehrlichia chaffeensis and E. ewingii are mainly reported from the United States and primarily transmitted by Amblyomma americanum ticks (Yabsley et al. 2011; Stoffel et al. 2014). Neorickettsia helminthoeca, the agent of salmon poisoning disease, is reported on the American continents and may infect dogs who ingest parasitized salmon (Headley et al. 2011). Lastly, the genera of Wolbachia is an obligate intracellular endosymbiont and likely mutualist living within numerous arthropod species and Filariidae nematodes (Taylor et al. 2012). The detection of Wolbachia species in blood of dog was associated with the presence of accompanying nematodes like D. immitis (Mehlhorn 2008).
In the management of canine vector‐borne diseases, the diagnosis should rely on the dog's clinical status as well as epidemiological information (Otranto et al. 2009). New Caledonia is an archipelago of Oceania, located in Melanesia (Pacific Ocean). The aim of this paper was to get an overview of the Anaplasmataceae and Filariidae species infecting dogs on this island and the association between Wolbachia spp. and Filariidae occurrence in dog blood samples. In this paper, we report the first description of A. platys, Wolbachia sp. endosymbiont of D. immitis associated with the presence of the nematode D. immitis, as well as Acanthocheilonema reconditum from the blood of dogs in New Caledonia.
Materials and methods
Sampling
In April 2009, blood from dogs living in kennels in New Caledonia was sampled in EDTA‐containing tubes by cephalic venipuncture. The kennels were at the Nouméa municipal animal shelter in the district of Ducos (South Province) and the Society for the Prevention of Cruelty to Animals (SPCA) in the district of Dumbéa. Military dogs were also sampled in Nandaïl, Népoui and Tontouta. Blood‐sucking parasites found on each sampled dog were removed. The animals were examined and sampled by a veterinarian after obtaining a verbal consent from people (owners) responsible of dogs. The dogs living in kennels were either stray dogs recovered by the relevant authorities or dogs abandoned/surrendered by their owners. No data are available about sanitary status or possible prophylaxis. After transport to the laboratory in Marseille, all samples were stored at −80°C.
Blood‐sucking parasite identification
Morphological identification was performed with a binocular microscope and carried out by an entomologist with a broad experience of arthropods of medical and veterinary importance. Blood‐sucking parasites were classified by family, genus and species using the available taxonomic keys and morphometric tables (Beaucournu & Launay 1990; Walker et al. 2000; Wall & Shearer 2008).
DNA extraction and PCR amplification
Total DNA was extracted from 200 μL of blood after digestion with proteinase K at +56°C for 16 h. DNA extraction was performed on the BioRobot EZ1 (Qiagen, Qiagen, Courtaboeuf, France) using a commercial DNA extraction kit (QIAamp DNA Mini Kit, Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. PCR screening and amplification of Anaplasmataceae species was conducted using primers and probes targeting the 23S rRNA gene followed by amplification and sequencing a 485‐base pair (bp) of the same gene as previously reported (Dahmani et al. 2017). For Filariidae species investigation, all DNA samples were firstly screened by a set of primers targeting the 421 bp‐long fragment of the mitochondrial 5S rRNA gene (Mourembou et al. 2015) and the 1551 bp‐long fragment of the mitochondrial cytochrome oxidase subunit (COI) gene using the set of primers specifically designed to amplify most filarial DNA. All probes and primers are listed in Table 1. All samples were then screened for the presence of D. immitis and Dirofilaria repens using the specific duplex qPCR as reported previously (Tahir et al. 2017).
Table 1.
Primers and probes used in this study
| Targeted microorganisms | Targeted sequences | Primers and probe | Sequences 5′‐3′ | Annealing temperature | References |
|---|---|---|---|---|---|
| qPCR | |||||
| Anaplasmataceae | 23S rRNA gene | TtAna‐F | ATAAGCTGCGGGGAATTGTC | 60°C | Dahmani et al. (2015a,2015b) |
| TtAna‐R | GTAACAGGTTCGGTCCTCCA | ||||
| TtAna‐S | FAM‐TGCAAAAGGTACGCTGTCAC‐TAMRA | ||||
| Conventional PCR | |||||
| Anaplasmataceae | 23S rRNA gene | Ana23S‐212f | GTTGAAAARACTGATGGTATGCA | 55°C | Dahmani et al. (2015a,2015b) |
| Ana23S‐753r | TGCAAAAGGTACGCTGTCAC | Mourembou et al. (2015) | |||
| Filariidae | 5S | S2 | GTTAAGCAACGTTGGGCCTGG | ||
| S16 | TTGACAGATCGGACGAGATG | ||||
| COI (PCR) | Filcox1‐F1 | TCCWGARATRGCGTTTCCTC | 48°C | This study | |
| Filcox1‐r1 | AACCATAGCCAACGCGACGAT | ||||
| COI (Sequencing) | Filcox1‐F1 | TCCWGARATRGCGTTTCCTC | |||
| Filcox1‐r1 | AACCATAGCCAACGCGACGAT | ||||
| Filcox1‐Fn | TTTTTGGACATCCTGARGTTT | ||||
| Oncox‐r1 | AATGAAAATGAGCYACAACAT | ||||
| Filcox1‐rn | ACCYTGTAWTCCAGCTAAAT | ||||
Sequencing and phylogenetic analyses
The amplicons were sequenced on a Biosystems 3130xl Genetic Analyzer (Thermo Fisher Scientific, France) using the DNA sequencing BigDye Terminator Kit (Perkin‐Elmer, Waltham, MA, USA) as described by the manufacturer. The sequences were assembled using the ChromasPro 1.7 (Technelysium Pty Ltd., Tewantin, Australia) and compared with Anaplasmataceae and Filariidae sequences available in the GenBank database using BLAST. Sequences obtained in this study were aligned with other ticks or Anaplasmataceae species sequences available on GenBank using CLUSTALW on Bioedit v3 (Hall 1999), and gaps and missing data were eliminated. Phylogenetic and molecular evolutionary analysis was inferred using the maximum likelihood method used on MEGA7 (Kumar et al. 2016), with the complete deletion option, based on the Hasegawa–Kishino–Yano (HYK) model for nucleotide sequences. A discrete gamma distribution was used to model evolutionary rate differences among sites. Initial trees for the heuristic search were automatically obtained by applying the Neighbour‐Joining and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. Statistical support for internal branches of the trees was evaluated by bootstrapping with 1000 iterations.
Results
A total of 64 dogs were sampled: 45 males and 19 females. Ages were available for 54 dogs. The average age was 4 years old [3 month–12 years]. In total, 51/64 (79.6, 95% CI: 69.8–89.5%) dogs were found to be infected by blood‐sucking parasites. In 44/64 (68.7–95% CI: 57.3–80.1%) dogs, 258 ticks were collected and identified as Rhipicephalus sanguineus s.l. Between four and eight ticks were removed from each dog. In addition, four Ctenocephalides felis fleas were removed from 4/64 (6.3, 95% CI: 0.3–18.1%) dogs and 24 lice Trichodectes canis were removed from 24/64 (37.5, 95% CI: 25.6–49.3%) dogs.
Screening using qPCR showed that 16/64 (25, 95% CI: 14.3–35.6%) samples contained DNA of bacteria belonging to the Anaplasmataceae family. After sequencing the 485 bp‐long amplicons of the 23S rRNA gene portion, BLAST analysis was conclusive for A. platys in eight samples, and Wolbachia sp. in eight samples. Two sequences of A. platys were obtained, respectively, from 6 to 2, then named, respectively, A. platys var1 and var2 (Fig. 1). The sequences of A. platys var1 are a 100% identity match with the A. platys amplified from dog blood reported in France (KM021412), Algeria (KM021427) and French Guiana (KM021414) (Dahmani et al. 2015a,2015b). Anaplasma platys var2 shared 99% identity with the A. platys var1 sequences cited above, and 100% homology with A. platys amplified from dogs in Algeria (KM021428) (Dahmani et al. 2015a,2015b). The total incidence was 12.5% (8/64, 95% CI: 4.4–20.6%). All infected dog were from a public (non‐military) shelter in Ducos, the incidence in this shelter was 8/25 (32, 95% CI: 13.7–50.2%).
Figure 1.

Phylogenetic tree showing the position of Anaplasma platys and Wolbachia sp. amplified from dog blood samples compared to other Anaplasmataceae bacteria available from GenBank. The evolutionary history was inferred using the maximum likelihood method based on the Hasegawa–Kishino–Yano model. A discrete Gamma distribution was used to model evolutionary rate differences among sites [4 categories (+G, parameter = 0.1479)]. The analysis involved 39 nucleotide sequences. All positions containing gaps and missing data were eliminated. There was a total of 434 positions in the final data set.
The 23S RNA gene sequences of Wolbachia were identical each other and show 96% homology with Wolbachia endosymbionts of Onchocerca ochengi when BLASTed. The 23S rRNA gene sequence of Wolbachia from D. immitis was not available in GenBank, so we have sequenced it from adult filaria obtained from a dog from French Guiana. When comparing the 23S sequences of Wolbachia from eight dogs from New Caledonia and adult D. immitis, we found a 100% identity match (Fig. 1).
The qPCRs specific to D. immitis and D. repens have identified 11/65 positive samples for only D. immitis. We obtained 480–583 bp‐long amplicons of filarial COI genes from nine of eleven dogs that tested positive for D. immitis by qPCR. The sequences obtained were identical to each to other and BLAST analysis showed 98% identity with the COI sequences of D. immitis reported worldwide (Fig. 2).
Figure 2.
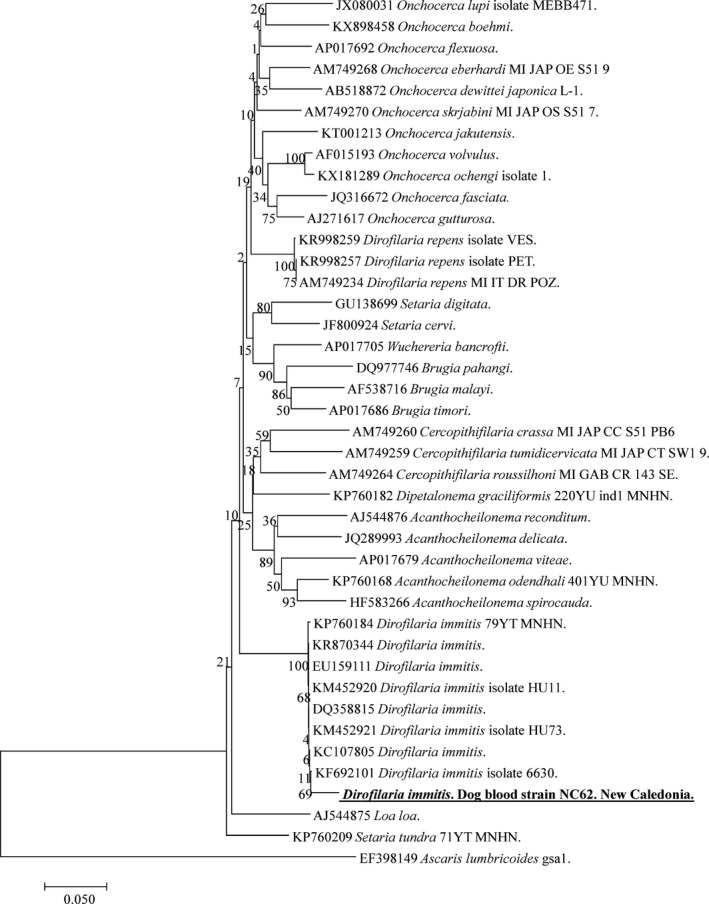
Phylogenetic tree showing the position of Dirofilaria immitis amplified from dog blood samples compared to other Filariidae nematodes available from GenBank. The evolutionary history was inferred using the maximum likelihood method based on the Hasegawa–Kishino–Yano model. A discrete Gamma distribution was used to model evolutionary rate differences among sites [4 categories (+G, parameter = 0.1479)]. The analysis involved 41 nucleotide sequences. All positions containing gaps and missing data were eliminated. There was a total of 526 positions in the final dataset.
We also screened the remaining 54 dogs that were negative for Dirofilaria‐specific qPCR. In two samples, we obtained a band in standard PCR targeting filarial COIs. We sequenced the portion of the amplicon and obtained sequences of 559 bp (identical to each other) showing 98% homology with the A. reconditum reported in Italy (JF461456 and AJ544876) (Fig. 3). For one sample positive for A. reconditum, we also amplified the 641 bp‐long sequence of the filarial 5S rRNA gene. When BLASTed, it showed 97% identity with Acanthocheilonema viteae (U31646), because, unfortunately, the 5S sequences belonging to A. reconditum were not available on GenBank.
Figure 3.

Phylogenetic tree showing the position of Acanthocheilonema reconditum amplified from dog blood samples compared to other Filariidae nematodes available from GenBank. The evolutionary history was inferred using the maximum likelihood method based on the Hasegawa–Kishino–Yano model. A discrete Gamma distribution was used to model evolutionary rate differences among sites [4 categories (+G, parameter = 0.2600)]. The analysis involved 42 nucleotide sequences. All positions containing gaps and missing data were eliminated. There was a total of 168 positions in the final dataset.
Co‐infection between Anaplasmataceae and Filariidae species were observed among the population of the dogs sampled in this study. Indeed, 7/8 samples positive for Wolbachia endosymbiont of D. immitis were infected with D. immitis, whereas the other four dogs infected with D. immitis did not reveal the presence of Wolbachia. One dog was found co‐infected both by A. reconditum and Wolbachia sp. which are usually associated with D. immitis. In addition, two cases of co‐infection were found: one of A. platys and D. immitis and another of A. platys and A. reconditum.
All the sequences obtained during this study were submitted to GenBank. For the Anaplasmataceae species: 23S rRNA gene for A. platys var1 and var2 as accession numbers KM021424 and KM021425, respectively. For Wolbachia sp. amplified from dog blood from New Caledonia, the accession number is KM021426, and for Wolbachia endosymbiont of D. immitis from French Guiana, KY347825. For the Filariidae species: the COI gene of D. immitis has the accession number KY347824, and A. reconditum has KY347823. For the 5S rRNA gene, the A. reconditum accession number is KY347822.
Discussion
This study reports the first detection of A. platys, D. immitis, and associated Wolbachia sp. as well as A. reconditum in dog blood samples from New Caledonia. Our results show that 12.5% (8/64) of dogs sampled in this study were infected with A. platys. In the Pacific region, the prevalence of A. platys in Australia was reported as 51, 36.2 and 32% in free‐roaming dogs tested from several remote Aboriginal communities (Brown et al. 2006; Barker et al. 2012; Hii et al. 2012), which is almost identical to our result of 32% in the Ducos shelter. In our study, all sampled military dogs (12/64) were found free of any Anaplasmataceae or Filariidae infection, as well as of free blood‐sucking parasites. All infected dogs were from the Dumbea municipal animal shelter or the Society for the Prevention of Cruelty to Animals (SPCA) (in the district of Dumbéa). Interestingly, in Australia, the prevalence of A. platys in dogs living with owners was 3.8% (Hii et al. 2015). In Italy, the vector‐borne disease was found to be significantly higher in a public shelter than in private kennels (Pennisi et al. 2012). So, it seems that the social status of the dogs and the quality of the treats they receive has an impact on the prevalence of vector‐borne diseases. In this study, 51/64 (68.8%) of the sampled dogs were found to be infested with blood‐sucking parasites. R. sanguineus s.l. were removed from 44/64 (68.7%) dogs, whereas, C. felis and T. canis were removed from 4/64 (6.25%) and 24/64 (37.5%) dogs, respectively. The vectors of A. platys seem to be R. sanguineus s.l. worldwide (Chomel 2011). All dogs infected with A. platys were found to be infested with R. sanguineus s.l. It appears that R. sanguineus s.l. are also major potential vectors of A. platys in New Caledonia. Furthermore, A. platys has previously been amplified from R. sanguineus s.l. and from the lice Heterodoxus spiniger in Australia. The possible role of the T. canis lice must be investigated in future studies.
The simultaneous detection of the Wolbachia endosymbiont of D. immitis and the associated nematode has been previously reported in Portugal and Spain (Maia et al. 2016). In our study, Wolbachia DNA was detected in seven of the 11 dogs infected with D. immitis. Dirofilaria immitis infections are widespread in both tropical and temperate regions throughout the world, including Australia where the climate is suitable for mosquito vectors of the genera Aedes, Culex and Anopheles (Smout et al. 2016). None of the tested dogs had D. repens infections. This species was reported ones in Australia in a patient who had been living in Sydney for over 40 years but made annual trips to Corfu, Greece (Stringfellow et al. 2002).
In this study, two dogs were found to be infected with A. reconditum. The 5S rRNA sequences obtained have 641 bp, whereas primers used in this study are expected to amplify a fragment of 421 bp. Indeed, the 5S rRNA is known to be variable in length and sequence, and only part of this gene and the spliced leader sequence SL1 are conserved in Brugia malayi, Brugia pahangi and D. immitis (Sanpool et al. 2016). Interesting, one of two dogs infected with A. reconditum was co‐infected with Wolbachia endosymbiont of D. immitis, however, D. immitis was not detected from this sample. Acanthocheilonema spp. are known to not harbour Wolbachia species (Maia et al. 2016). It is possible that this dog was co‐infected with both A. reconditum and D. immitis, although specific qPCR for D. immitis was negative. Co‐infection with two Filariidae species has been previously reported, including A. drancunculoides in co‐infection with D. immitis in Portugal (Maia et al. 2016). This parasite is vectored by fleas (C. canis, C. felis, Pulex irritans, P. simulans, Echidnophaga gallinae) and lice (Heterodoxus spiniger, Linognathus setosus) (Otranto et al. 2013). In total, 4/64 (6.25%) and 24/64 (37.5%) dogs were found to be infested by C. felis fleas and T. canis lice, respectively, in this study. Both dogs infected with A. reconditum in our study were found to be infested by R. sanguineous s.l but not by fleas or lice. However, the role of R. sanguineous s.l in the transmission of A. reconditum has never been shown.
One dog was found to be co‐infected by A. reconditum and A. platys. Co‐infection between A. platys and Rickettsiae or other tick‐borne pathogens occurs frequently in dogs (Andersson et al. 2013). The increased co‐infection by multiple vector‐borne diseases and other agents in dogs is usually noted in endemic areas where the prevalence of both organisms is high, and has an important effect on disease expression and the ability to treat patients and animals effectively (Nicholson et al. 2010).
In conclusion, this study reports the first description of A. platys, D. immitis and associated Wolbachia, as well as A. reconditum in dogs sampled in New Caledonia. This study opens the way for further investigation of Anaplasmataceae and Filariidae species infecting dogs as little is known about species occurrences and potentially associated vectors.
Source of funding
This study was supported by the AMIDEX project (n° ANR‐11‐IDEX‐0001‐02) funded by the “Investissements d'Avenir” French Government programme, managed by the French National Research Agency (ANR) and Foundation Méditerranée Infection (http://www.mediterranee-infection.com). The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
All of the authors declare no conflict of interest related to this article.
Ethics statement
The authors declare that all institutional guidelines for use of dogs sampled in this study were followed.
Contributions
DR, BD, OM and FF designed the study; BD and OC collected the samples; MD and DT conducted the lab experiments; MD, DT and OM analysed the data; MD, BD and OM prepared the manuscript.
References
- Andersson M., Turcitu M.A., Stefanache M., Tamba P., Barbuceanu F. & Chitimia L. (2013) First evidence of Anaplasma platys and Hepatozoon canis co‐infection in a dog from Romania – A case report. Ticks and Tick‐borne Diseases 4, 317–319. [DOI] [PubMed] [Google Scholar]
- Barker E.N., Langton D.A., Helps C.R., Brown G., Malik R., Shaw S.E. & Tasker S. (2012) Haemoparasites of free‐roaming dogs associated with several remote Aboriginal communities in Australia. BMC Veterinary Research 8, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaucournu J.C., Launay H. (1990). Les Puces (Siphonaptera) de France et du Bassin méditerranéen occidental. Faune de France 76. 548.
- Brown G.K., Canfield P.J., Dunstan R.H., Roberts T.K., Martin A.R., Brown C.S. & Irving R. (2006) Detection of Anaplasma platys and Babesia canis vogeli and their impact on platelet numbers in free‐roaming dogs associated with remote aboriginal communities in Australia. Australian Veterinary Journal 84, 321–325. [DOI] [PubMed] [Google Scholar]
- Chomel B. (2011) Tick‐borne infections in dogs‐an emerging infectious threat. Veterinary Parasitology 179, 294–301. [DOI] [PubMed] [Google Scholar]
- Dahmani M., Loudahi A., Mediannikov O., Fenollar F., Raoult D. & Davoust B. (2015a) Molecular detection of Anaplasma platys and Ehrlichia canis in dogs from Kabylie, Algeria. Ticks and Tick‐borne Diseases 6, 198–203. [DOI] [PubMed] [Google Scholar]
- Dahmani M., Marié J.L., Mediannikov O., Raoult D. & Davoust B. (2015b) First identification of Anaplasma platys in the blood of dogs from French Guiana. Vector‐Borne Zoonotic Diseases. 15, 170–172. [DOI] [PubMed] [Google Scholar]
- Dahmani M., Davoust B., Tahir D., Raoult D., Fenollar F. & Mediannikov O. (2017) Molecular investigation and phylogeny of Anaplasmataceae species infecting domestic animals and ticks in Corsica, France. Parasites and Vectors 10, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumler J.S., Barbet A.F., Bekker C.P., Dasch G.A., Palmer G.H., Ray S.C. et al (2001) Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila . International Journal of Systematic and Evolutionary Microbiology 51, 2145–2165. [DOI] [PubMed] [Google Scholar]
- Dumler J.S., Choi K., Garcia‐Garcia J.C., Barat N.S., Scorpio D.G., Garyu J.W. et al (2005) Human granulocytic anaplasmosis and Anaplasma phagocytophilum . Emerging Infectious Diseases 11, 1828–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: a user‐friendly biological sequences alignment editors and analysis program for Windows 95/98/NT. 95–98, Nucleic Acids Symposium Series No. 41.
- Harrus S. & Waner T. (2011) Diagnosis of canine monocytotropic ehrlichiosis (Ehrlichia canis): an overview. The Veterinary Journal 187, 292–296. [DOI] [PubMed] [Google Scholar]
- Headley S.A., Scorpio D.G., Vidotto O. & Dumler J.S. (2011) Neorickettsia helminthoeca and salmon poisoning disease: a review. The Veterinary Journal 187, 165–173. [DOI] [PubMed] [Google Scholar]
- Hii S.F., Kopp S.R., Thompson M.F., O'Leary C.A., Rees R.L. & Traub R.J. (2012) Canine vector‐borne disease pathogens in dogs from south‐east Queensland and north‐east Northern Territory. Australian Veterinary Journal 90, 130–135. [DOI] [PubMed] [Google Scholar]
- Hii S., Traub R., Thompson M., Henning J., O'Leary C., Burleigh A. et al (2015) Canine tick‐borne pathogens and associated risk factors in dogs presenting with and without clinical signs consistent with tick‐borne diseases in northern Australia. Australian Veterinary Journal 93, 58–66. [DOI] [PubMed] [Google Scholar]
- Kohn B., Silaghi C., Galke D., Arndt G. & Pfister K. (2011) Infections with Anaplasma phagocytophilum in dogs in Germany. Research in Veterinary Science 91, 71–76. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G. & Tamura K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia C., Altet L., Serrano L., Cristóvão J.M., Tabar M.D., Francino O. et al (2016) Molecular detection of Leishmania infantum, filariae and Wolbachia spp. in dogs from southern Portugal. Parasites & Vectors 9, 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlhorn H. (2008) Filariidae. In: Encyclopedia of Parasitology Springer Berlin Heidelberg; pp. 522–522. [Google Scholar]
- Mourembou G., Fenollar F., Lekana‐Douki J.B., Ndjoyi Mbiguino A., Maghendji Nzondo S., Matsiegui P.B. et al (2015) Mansonella, including a potential new species, as common parasites in children in Gabon. PLoS Neglected Tropical Diseases 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W.L., Allen K.E., Mcquiston J.H., Breitschwerdt E.B. & Little S.E. (2010) The increasing recognition of rickettsial pathogens in dogs and people. Trends in Parasitology 26, 205–212. [DOI] [PubMed] [Google Scholar]
- Otranto D., Dantas‐Torres F. & Breitschwerdt E.B. (2009) Managing canine vector‐borne diseases of zoonotic concern: part two. Trends in Parasitology 25, 228–235. [DOI] [PubMed] [Google Scholar]
- Otranto D., Dantas‐Torres F., Brianti E., Traversa D., Petrić D., Genchi C. & Capelli G. (2013) Vector‐borne helminths of dogs and humans in Europe. Parasites & Vectors 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi M.G., Caprì A., Solano‐Gallego L., Lombardo G., Torina A. & Masucci M. (2012) Prevalence of antibodies against Rickettsia conorii, Babesia canis, Ehrlichia canis, and Anaplasma phagocytophilum antigens in dogs from the Stretto di Messina area (Italy). Ticks and Tick‐borne Diseases 3, 315–318. [DOI] [PubMed] [Google Scholar]
- Sanpool O., Tantrawatpan C., Thanchomnang T., Janwan P., Intapan P.M., Rodpai R. et al (2016) Pyrosequencing using SL and 5S rRNA as molecular markers for identifying zoonotic filarial nematodes in blood samples and mosquitoes. Vector‐Borne Zoonotic Dis. 16(5), 326–333. [DOI] [PubMed] [Google Scholar]
- Smout F.A., Skerratt L.F., Butler J.R.A., Johnson C.N. & Congdon B.C. (2016) Dingoes (Canis dingo Meyer, 1793) continue to be an important reservoir host of Dirofilaria immitis in low density housing areas in Australia. Veterinary Parasitology 215, 6–10. [DOI] [PubMed] [Google Scholar]
- Stoffel R.T., McClure J.C., Butcher M.M., Johnson G.C., Roland W., Cheng C. et al (2014) Experimental infection of Rhipicephalus sanguineus with Ehrlichia chaffeensis . Veterinary Microbiology 172, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringfellow G.J., Francis I.C., Coroneo M.T. & Walker J. (2002) Orbital dirofilariasis. Journal of Clinical and Experimental Ophthalmology 30, 378–380. [DOI] [PubMed] [Google Scholar]
- Tahir D., Bittar F., Barré‐Cardi H., Sow D., Dahmani M., Mediannikov O. et al (2017) Molecular survey of Dirofilaria immitis and Dirofilaria repens by new real‐time TaqMan® PCR assay in dogs and mosquitoes (Diptera: Culicidae) in Corsica (France). Veterinary Parasitology 235, 1–7. [DOI] [PubMed] [Google Scholar]
- Taylor M., Mediannikov O., Raoult D. & Greub G. (2012) Endosymbiotic bacteria associated with nematodes, ticks and amoebae. FEMS Immunology and Medical Microbiology 64, 21–31. [DOI] [PubMed] [Google Scholar]
- Walker J.B., Keirans J.E. & Horak I.G. (2000) The genus Rhipicephalus (Acari, Ixodidae): a guide to the brown ticks of the world. Tropical Animal Health and Production 32, 417–418. [Google Scholar]
- Wall R. & Shearer D. (2008) Veterinary ectoparasites: biology, pathology and control. 2nd edn John Wiley & Sons: Oxford. [Google Scholar]
- Yabsley M.J., Adams D.S., Connor T.P.O., Chandrashekar R. & Little S.E. (2011) Experimental primary and secondary infections of domestic dogs with Ehrlichia ewingii . Veterinary Microbiology 2, 315–321. [DOI] [PubMed] [Google Scholar]


