Abstract
Inflammatory genes serve a crucial role in the pathogenesis of inflammation-associated tumors. However, as recent studies have mainly focused on the effects of single inflammatory genes on colorectal cancer (CRC), but not on the global interactions between genes, the underlying mechanisms between inflammatory genes and CRC remain unclear. In the current study, two inflammation-associated networks were constructed based on inflammatory genes, differentially expressed genes (DEGs) in CRC vs. normal samples, and protein-protein interactions (PPIs). These networks included an inflammation-related neighbor network (IRNN) and an inflammation-related DEG network (IRDN). Notably, the results indicated that the inflammatory genes served as important CRC-associated genes in the IRNN. Certain inflammatory genes were more likely to be network hubs and exhibited higher betweenness centralities, indicating that these inflammatory hub genes had central roles in the communication between genes in the IRNN. By contrast, in the IRDN, functional enrichment analysis revealed that genes were enriched in numerous cancer-associated functions and pathways. Subsequently, 14 genes in a module were identified in the IRDN as the potential biomarkers associated with disease-free survival (DFS) in CRC patients in the GSE24550 dataset, the prognosis of which was further validated using three independent datasets (GSE24549, GSE34551 and GSE103479). All 14 genes (including BCAR1, CRK, FYN, GRB2, LCP2, PIK3R1, PLCG1, PTK2, PTPN11, PTPN6, SHC1, SOS1, SRC and SYK) in this module were inflammatory genes, emphasizing the critical role of inflammation in CRC. In conclusion, these findings based on integrated inflammation-associated networks provided a novel insight that may help elucidate the inflammation-mediated mechanisms involved in CRC.
Keywords: colorectal cancer, inflammatory genes, regulatory network, survival analysis, biomarkers
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer among males and females (1). Surgical resection is an effective treatment for early-stage CRC, while chemotherapy is the main treatment for patients with advanced CRC. In recent years, effective observation strategies following curative treatment for CRC have helped improve the overall survival (OS) of patients, and a number of CRC patients are eligible for such curative treatment (2–4). The fecal immunochemical test (FIT), a simple and easy method, has been widely used in CRC screening programs (5–7). However, the 5-year survival rate significantly declines when cancer cells spread to adjacent organs or lymph nodes (8–10). Due to the genetic heterogeneity of CRC, studying the tumor initiation and progression at a molecular level can help to uncover the pathogenesis of CRC. During the past decades, researchers have identified various important gene biomarkers, including tumor oncogenes (such as KRAS, BRAF and PIK3CA) and suppressor genes (such as A00PC, TP53 and PTEN). These genes are critical for the genesis and development of CRC, demonstrating the potential clinical significance for the treatment of CRC (11,12).
As accumulating evidence has suggested inflammation is a tumor-promoting hallmark that contributes to tumor initiation and progression (13). CRC has been reported to be associated with chronic bowel inflammation (14–16), indicating the crucial roles of inflammatory genes in the tumorigenesis and development of CRC. Additionally, a number of studies have reported the importance of single inflammatory genes in CRC. For instance, Cho et al (17) revealed that genetic variation in PPARGC1A affected the role of diet-associated inflammation in CRC. Furthermore, Li et al (18) identified that the inflammatory molecule PSGL-1, involved in the nuclear factor-κB signaling pathway, can promote CRC growth by activating macrophages. However, the study of inflammatory genes in the context of functional interaction networks remains unclear, which drives us to predict novel network-based inflammation-associated signatures. In addition, a variety of public gene expression profiling datasets and protein-protein interactions (PPIs) make it possible to study the network of inflammatory genes further in order to identify novel biomarkers for CRC. Chuang et al (19) used a protein-network-based approach to identify biomarkers as sub-networks. Similarly, by developing a network-based method, Li et al (20) identified cancer prognostic biomarkers based on microarray and network datasets.
The present study aimed to investigate how inflammatory genes functioned in the context of inflammation-associated networks and their prognostic value in CRC, and two inflammation-associated networks were constructed. Topological properties were used to measure the network structure, as well as the crucial positions of the inflammatory and inflammation-associated genes in the two networks in CRC. Furthermore, a 14-gene module was identified in the IRDN, and notably, all 14 genes in this module were inflammatory genes. The prognostic significance of this module comprising 14 inflammatory genes was validated by three independent datasets. The current study findings highlighted the novel role of the inflammation-associated network in CRC, providing an insight into the inflammation-mediated mechanisms involved in this disease.
Materials and methods
Data resources
CRC patients with whole-genome gene expression profiles (Affymetrix Human Exon 1.0 ST Array) were downloaded from the publicly available Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). A total of 90 specimens were included in the study of network construction, including 77 tumor and 13 normal samples from the study by Sveen et al (21) with the accession number GSE24550 (Table I). In addition, another three GEO datasets, namely GSE24549, GSE24551 and GSE103479 (Table I) (22,23), were downloaded for validation analyses. The GSE24550 dataset, which is a subset of GSE24551, was selected as a training set. Furthermore, CRC patients' stage information was obtained for these four datasets and other clinical features (age, gender, BRAF/KRAS/P53 mutations) for the GSE103479 dataset.
Table I.
Clinical features of all CRC patients included in this study.
| Characteristic | GSE24550 (n=77) | GSE24549 (n=83) | GSE24551 (n=160) | GSE102479 (n=155) |
|---|---|---|---|---|
| Stage, no. (%) | ||||
| II | 44 (57.1) | 46 (55.4) | 90 (56.2) | 83 (53.5) |
| III | 33 (42.9) | 37 (44.6) | 70 (43.8) | 72 (46.5) |
| Age, no. (%) | ||||
| ≤65 | n/a | n/a | n/a | 51 (32.9) |
| >65 | n/a | n/a | n/a | 102 (56.8) |
| Gender, no. (%) | ||||
| Male | n/a | n/a | n/a | 87 (56.1) |
| Female | n/a | n/a | n/a | 68 (43.9) |
| BRAF mutation | ||||
| WT | n/a | n/a | n/a | 122 (78.7) |
| MT | n/a | n/a | n/a | 15 (9.7) |
| KRAS mutation | ||||
| WT | n/a | n/a | n/a | 83 (53.5) |
| MT | n/a | n/a | n/a | 54 (34.8) |
| P53 mutation | ||||
| WT | n/a | n/a | n/a | 59 (38.1) |
| MT | n/a | n/a | n/a | 78 (50.3) |
WT, wild type; MT, mutation type; n/a, not available.
Gene Ontology (GO) (24) terms associated with the inflammatory response were obtained from the study by Plaisier et al (25). The genes annotated to these inflammation-associated GO terms were obtained from the AmiGO2 tool (26) of the GO Consortium. Finally, 909 inflammatory genes were collected for subsequent analyses.
The protein-protein interaction (PPI) data were downloaded from the Human Protein Reference Database (HPRD) release 9 (http://www.hprd.org/) (27). It contained >42,000 manually curated interactions between 9,826 human genes.
Differentially expressed gene (DEG) analysis
The raw array data (.CEL files) of samples in the four GEO datasets were uniformly pre-processed using the Robust Multichip Average algorithm for background correction, quantified normalization and log2-transformation (28). To account for the heterogeneity of multiple microarray datasets in systematic measurements, each dataset was standardized independently by the Z-score transformation to balance the expression intensities of each probe (29). DEGs were identified using a two-tailed t-test for GSE24550. Genes with a false discovery rate (FDR) cutoff value of 0.001 following adjustment of the P-value were considered as DEGs. The unsupervised hierarchical clustering of CRC samples and DEGs was performed with R software (https://www.r-project.org/) using the Euclidean distance and complete linkage method.
Network construction and analysis
Cytoscape version 3.2.0 (30) was used for the construction of the networks in the current study. The first network was established according to the following procedure: The human PPI network was initially downloaded from the HPRD, all the inflammatory gene symbols were acquired from AmiGO2, and all the inflammatory genes were mapped to the human PPI network. Subsequently, a sub-network including inflammatory genes and their direct interacting genes in the network (referred to as inflammatory neighbor genes) was selected. Finally, the sub-network was termed the inflammation-related neighbor network (IRNN). The second sub-network was constructed as follows: DEGs of CRC were mapped to IRNN, and then a sub-network including DEGs and their direct interacting genes in the IRNN (referred to as DEG neighbor genes) was selected. The second sub-network was termed the inflammation-related driver network (IRDN) for subsequent analyses.
In order to explore the topological properties of the inflammatory genes in the IRNN and IRDN, the following features were analyzed: Degree, betweenness centrality (BC) and closeness centrality (CC), which were used to decipher the structure of the sub-networks and to identify specific vital molecules. Degree was used to determine the number of neighbors for each node. BC represented the key role of a node in communication and information diffusion (31). Node CC measured the local cohesiveness, such as how close a node is to other nodes. The IRNN and IRDN visualization and topological properties were analyzed using Cytoscape version 3.2.0 and the R software.
Identifying prognostic modules in IRDN
The cFinder algorithm (32) is a classic method used to identify modules from a network, in which the identification and visualization of overlapping dense groups of nodes was conducted using the Clique Percolation Method (33). Modules were identified through this method in the present study by locating the k-clique percolation clusters of the network. A k-clique percolation cluster includes the following two parts: i) all nodes that can be reached via chains of adjacent k-cliques from each other; and ii) the links in these cliques. Larger values of k correspond to a higher stringency during the identification of dense groups and provide smaller groups with a higher density of links inside them.
Functional enrichment analysis
Genes were functionally annotated to identify enriched pathways based on DAVID Bioinformatics Resources (version 6.7; http://david.abcc.ncifcrf.gov/) (34). The DAVID enrichment analyses were limited to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and GO-FAT biological process (BP) terms, with the whole human genome as background. Functional categories were visualized and clustered using the Enrichment Map plugin (35) in Cytoscape version 3.2.0.
Survival analysis
In order to validate whether the module identified in the aforementioned step was associated with CRC patient survival, the expression of mRNAs in the module was extracted. Next, Cox proportional hazard analysis was used to obtain the regression coefficient of each gene associated with patient survival. The classifier was built as the linear combination of the gene expression values of the selected genes using the standardized Cox regression coefficient as the weight. A risk score formula for each patient was established by including the expression values of each selected gene weighted by their estimated regression coefficients in the univariate Cox regression analysis. As a result, patients were divided into the high-risk and low-risk groups (36) using the median value of the risk score as the threshold. Kaplan-Meier survival plots and log-rank tests were used to assess the differences in disease-free survivalb (DFS) duration between the high-risk and low-risk patients. In addition, the sensitivity and specificity of the module for survival prediction was evaluated using receiver operating characteristic (ROC) curve analysis, and the area under the ROC curve (AUC) was calculated.
Statistical analysis
In the construction of IRNN, two-tailed t-test and FDR adjustion were used to identify CRC-related DEGs. Furthermore, univariate and multivariate Cox regression analyses and log-rank test were applied in survival analysis.
Results
Inflammatory genes serve a crucial role in CRC
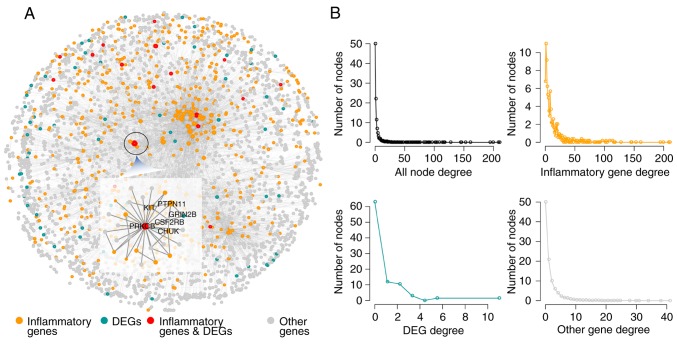
Based on the PPI network, an IRNN was constructed, which included 9,293 interactions of 3,526 nodes (575 inflammatory genes and their 2,928 neighbors; Fig. 1A). In the IRNN, there were 17 inflammatory genes (such as CCL19, CD36, COL1A1, CXCL13, ITK and KIT) that were DEGs, as well as 558 inflammatory genes (including PRKACA, TRAF2, PIK3R1, SHC1, PTPN11, LCK and GRB2) that were associated with the DEGs.
Figure 1.
Properties of the IRNN. (A) The global IRNN and (B) the degree distribution of the nodes in the IRNN are shown. IRNN, inflammation-related neighbor network; DEG, differentially expressed gene.
In order to explore the construction and features of the IRNN, network topology analysis was performed. As observed, the global degree distribution of nodes in the IRNN closely followed the power law distribution (Fig. 1B). Furthermore, the inflammatory genes, DEGs and other nodes exhibited a small-world network organization (Fig. 1B), which suggested that the IRNN constructed was biologically significant.
Inflammatory genes serve critical hub roles by interacting with DEGs in CRC
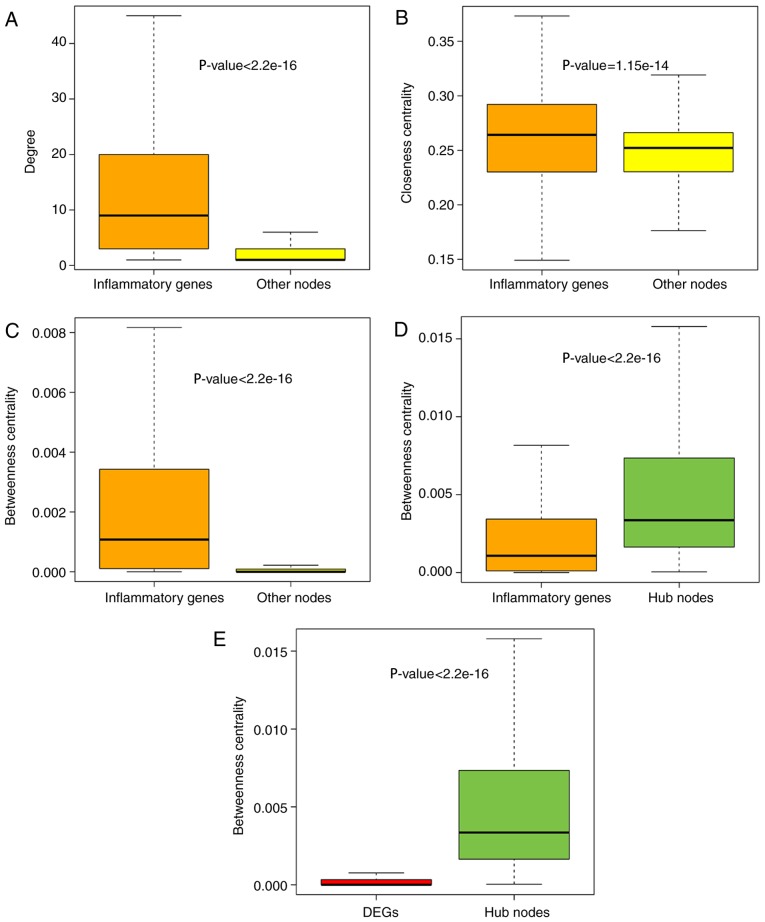
To further explore the role of inflammatory genes in the IRNN, analysis of the topological features, including the degree, CCs and BCs, was performed. The results demonstrated that inflammatory genes in the IRNN had a higher degree, CCs and BCs compared with their neighbors that were non-inflammatory genes in the IRNN (P<2.2×10−16 for degree; P=1.15×10−14 for CC; P<2.2×10−16 for BC; Wilcoxon rank sum test; Fig. 2A–C, respectively), indicating that inflammatory genes were central within the IRNN.
Figure 2.
Comparison analysis of topological properties of inflammatory genes and other mRNAs in the IRNN. Box plot of the (A) degree, (B) BC and (C) CC of inflammatory genes and other nodes in the IRNN. (D) Box plot of BC of hub nodes and inflammatory genes in IRNN. (E) Box plot of BC of hub nodes and DEGs in IRNN. IRNN, inflammation-related neighbor network; DEG, differentially expressed gene; BC, betweenness centrality; CC, closeness centrality.
As observed earlier, inflammatory genes tended to be located in the hub node positions in the IRNN; therefore, the present study further deciphered these inflammatory hub genes in detail. Previously, hubs were typically defined as the top 10–20% of nodes in the networks based on their degree (37,38). Thus, the top 10% of nodes were selected as the hub components based on the highest degrees, identifying a total of 347 hub nodes. Furthermore, all the hub nodes were found to belong to inflammatory genes with a significantly higher BC compared with that of all the inflammatory genes in the IRNN (P<2.2×10−16; Fig. 2D). These findings indicated that certain inflammatory genes had critical hub roles in the IRNN. However, the BCs of hub genes were higher in comparison with those of DEGs (P<2.2×10−16; Fig. 2E), indicating that these DEGs served critical roles in the communication with their neighboring nodes in the IRNN. The KIT gene, a DEG in the IRNN sub-network, encodes c-kit tyrosine kinase whose inhibitor STI571 reportedly exhibits a substantial therapeutic activity in patients with CRC (39). KIT, which interacted with two hub inflammatory genes (FYN and SRC) in the IRNN, is inhibited by dasatinib, which is studied in solid tumors such as CRC by combining with capecitabine and oxaliplatin (40). These observations demonstrated that hub inflammatory gene-associated DEGs were more likely to be essential for CRC development and progression.
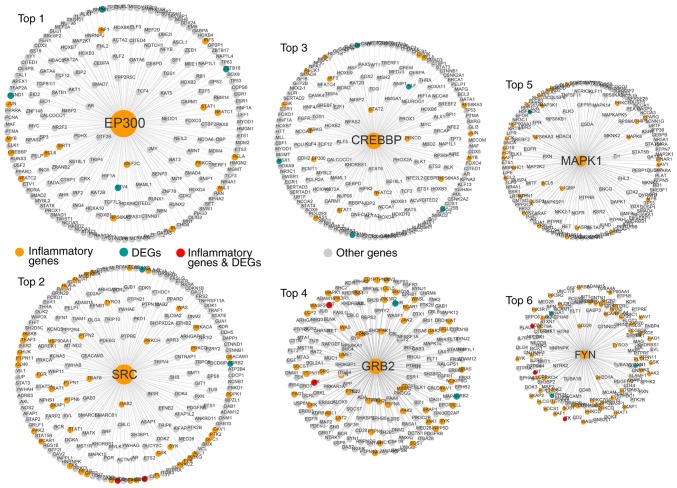
The top six inflammatory genes (EP300, SRC, CREBBP, GRB2, MAPK1 and FYN) in terms of the node degree had a direct connection in the network, indicating that inflammatory genes function as hubs in the sub-networks (Fig. 3).
Figure 3.
Top six inflammatory genes ranked in terms of the gene degree, including EP300, SRC, CREBBP, GRB2, MAPK1 and FYN. DEG, differentially expressed gene.
Inflammatory genes directly interact with CRC-associated DEGs
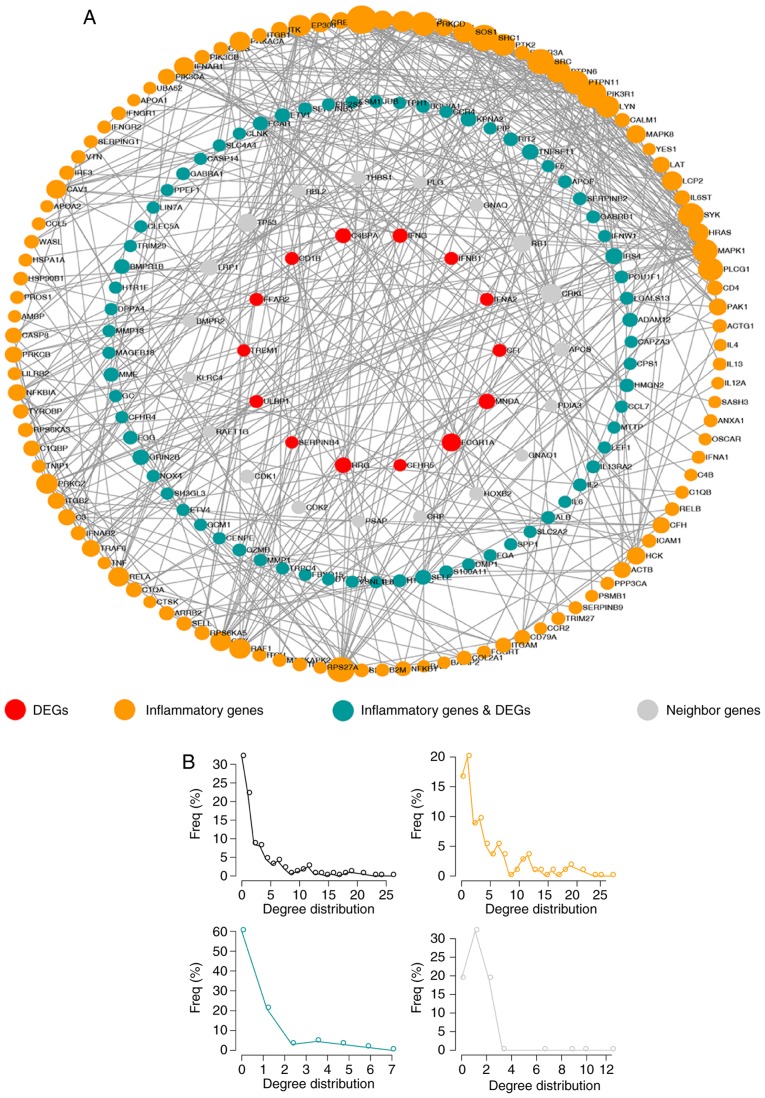
To further explore the association between inflammatory genes and DEGs, a network termed IRDN was constructed, which included DEGs and their neighbors extracted from the IRNN (Fig. 4A). The IRDN included 209 genes, of which 83 were DEGs. In total, 121 of the genes in IRDN were inflammatory genes, and 17 of these were also DEGs. Similar to the nodes in IRNN, the nodes in the IRDN also exhibited a small-world network organization (Fig. 4B), which suggested that the IRDN was also biologically significant. These results suggested that the IRDN obtained from IRNN may serve a crucial role in the biogenesis of CRC.
Figure 4.
Properties of the IRDN. (A) The global IRDN is shown, and the size of the sphere represents the degree of the gene. (B) The degree distribution of mRNAs in the IRDN. IRDN, inflammation-related DEG network; DEG, differentially expressed gene.
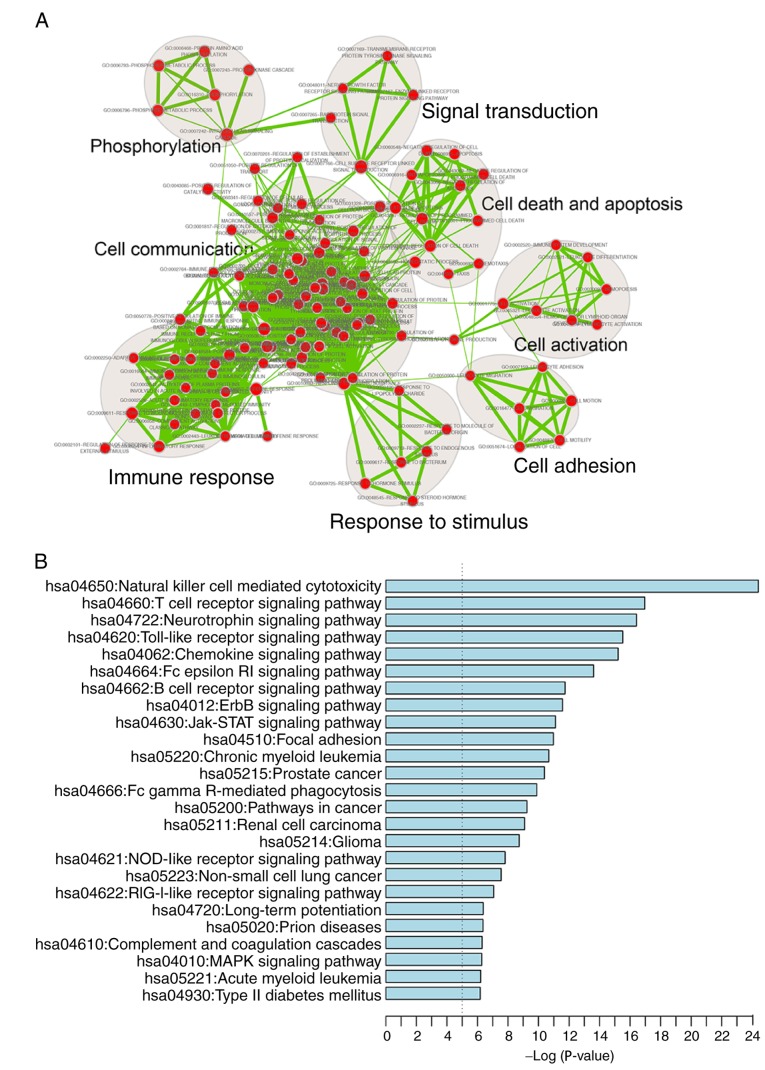
A functional enrichment analysis of all genes in IRDN was also performed based on GO and KEGG pathway analyses. IRDN mRNAs were enriched in 200 GO_BP_FAT terms (FDR<0.001) mainly in eight functional clusters, including cell adhesion, cell communication, cell activation, immune response, cell death and apoptosis, signal transduction, response to stimulus and phosphorylation (Fig. 5A). In addition, 25 KEGG pathways (FDR<0.01) were involved, including cancer, focal adhesion and several signaling pathways (Fig. 5B). All enriched signaling pathways, including ErbB, Jak-STAT, MAPK and NOD-like receptor signaling, are known to be contributors to CRC pathogenesis (41–44).
Figure 5.
(A) Functional enrichment map of GO_BP_FAT terms. Each node represented a GO term, which was grouped and annotated by GO similarity. Each edge represented whether there was a shared gene between two GO terms. Node size represented the number of genes in the GO term. (B) Significantly enriched KEGG pathways of mRNAs in the IRDN. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; IRDN, inflammation-related DEG network; DEG, differentially expressed gene.
Identifying inflammatory network-based biomarkers in CRC
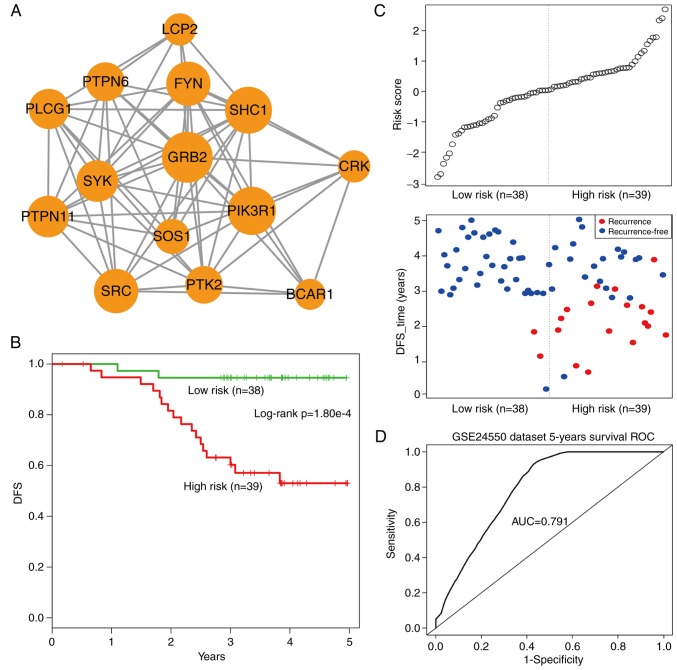
Using the cFindeR software, two inflammation-associated modules in IRDN with k-clique ≥5 were identified. Next, the study investigated whether these two inflammatory gene-associated modules have a prognostic potential in CRC. Using the training dataset GSE24550 (Table II), a risk-score formula was created according to the expression of these two module genes to generate DFS prediction. Using the median risk score of the training series as the cutoff point, the risk scores of the module genes were calculated for each patient and then patients were ranked according to their risk score. The patients were grouped into the high-risk or low-risk categories using the median risk score of the training series as the cutoff point. As a result, the module comprising 14 inflammatory genes with k-clique=6 (Fig. 6A) was able to divide patients into the high- and low-risk groups with a significantly reduced DFS observed in the high-risk group (log-rank test, P=1.80×10−4; Fig. 6B). Additionally, the distribution of gene risk score and the survival status of the training dataset are shown in Fig. 6C. Furthermore, a time-dependent ROC curve analysis was performed to evaluate the sensitivity and specificity for module survival prediction. The module achieved an AUC value of 0.791 (Fig. 6D), suggesting a substantially effective performance.
Table II.
Gene Expression Omnibus microarray datasets used in the present study.
| Datasets | Platform | Number of patients | Overall type | No. of tumor samples | No. of normal samples |
|---|---|---|---|---|---|
| GSE24550 | HuEx-1_0-st | 90 | DFS | 77 | 13 |
| GSE24549 | HuEx-1_0-st | 83 | DFS | 83 | 0 |
| GSE24551 | HuEx-1_0-st | 173 | DFS | 160 | 13 |
| GSE103497 | ADXECv1a520743 | 156 | OS | 156 | 0 |
DFS, disease-free survival; OS, overall survival.
Figure 6.
Overview of 14-inflammatory-gene-module and its prognostic ability in assessing the clinical outcome of the training set colorectal cancer patients. (A) Overview of 14-inflammatory-gene-module, with the size of the sphere depicting the degree of the gene. (B) Kaplan-Meier survival curves for disease-free survival of patients in the training set GSE24550 with high- and low-risk scores. The P-value was calculated using the log-rank test. (C) Risk score analysis of the module in GSE24550. (D) ROC curve analysis and the AUC indicating the sensitivity and specificity of the module for survival prediction in the GSE24550 dataset. DFS, disease-free survival; ROC, receiver operating characteristic; AUC, area under the ROC curve.
Validating the prognostic potential of 14-inflammatory-gene-module in independent datasets
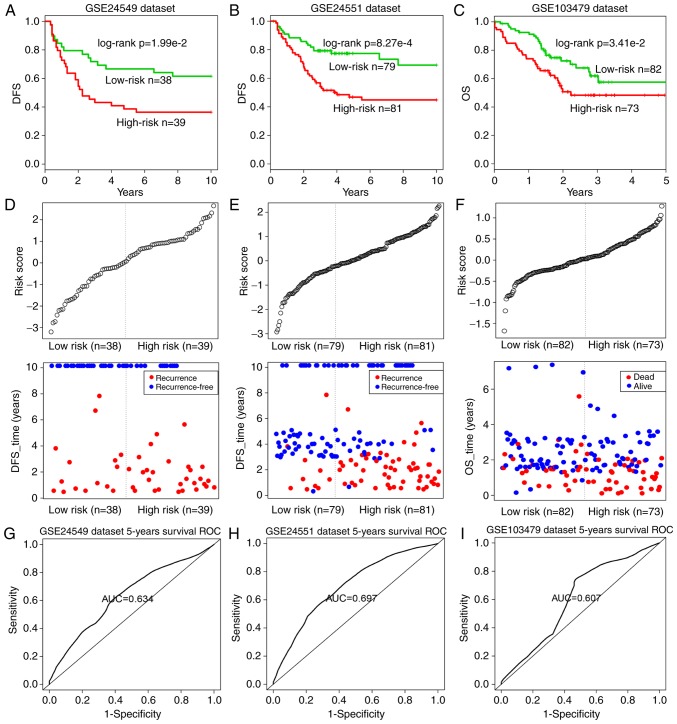
To validate the prognostic performance of the 14-inflammatory-gene-module, three independent GEO datasets were used in the subsequent analyses (Table II). Using the same risk score formula as earlier, patients in each dataset were classified into the high-risk and low-risk groups using the median score of the training series as the cutoff point. Consistent with the previous findings, patients in the high-risk group had significantly shorter median DFS or OS when compared with those in the low-risk group (GSE24549 patients: Log-rank test P=1.99×10−2, Fig. 7A; GSE24551 patients: Log-rank test P=8.27×10−4, Fig. 7B; GSE103479 patients: Log-rank test P=3.41×10−2, Fig. 7C). Similarly, the distribution of gene risk scores and the survival statuses of the three datasets are displayed in Fig. 7D–F. Patients with high-risk scores tended to present poorer clinical outcomes compared with patients with low-risk scores. The time-dependent ROC curve analysis was performed to evaluate the sensitivity and specificity for module survival prediction in these three GEO datasets. The module achieved AUC values of 0.634, 0.697 and 0.607 in the GSE24549, GSE24551 and GSE103479 datasets, respectively, indicating that high prediction performance (Fig. 7G–I).
Figure 7.
Survival curves and risk score analyses of the 14-inflammatory-gene-module in three independent datasets. Kaplan-Meier survival curves for DFS of patients with high- and low-risk scores in the (A) GSE24549, (B) GSE24551 and (C) GSE103497 datasets. Log-rank test was used to calculate the P-values. Risk score analysis of the module in the patients of the (D) GSE24549, (E) GSE24551 and (F) GSE103497 datasets. ROC curve analysis and AUC value indicating the sensitivity and specificity of the module for survival prediction in the (G) GSE24549, (H) GSE24551 and (I) GSE103497 datasets. DFS, disease-free survival; ROC, receiver operating characteristic; AUC, area under the ROC curve.
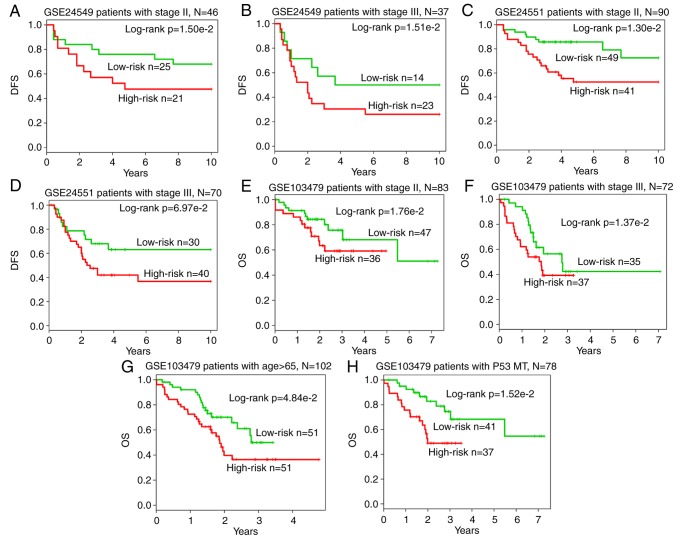
Subsequent to further adjusting for other clinical markers univariate analysis indicated that the 14-inflammatory-gene-module was significantly associated with the DFS of CRC patients from the GSE24549 [hazard ratio (HR)=1.28; 95% confidence interval (CI)=0.99–1.66; P=0.052] and GSE24551 datasets (HR=1.88; 95% CI=1.42–2.47; P<0.001), as well as the OS of patients from the GSE103479 dataset (HR=2.13; 95% CI=1.24–3.64; P=0.006), as an independent risk factor (Table III). Subsequently, when multivariate analysis was performed to investigate the independence of the module to other clinical factors, the module was not independent of the stage in GSE24549 (HR=1.93; 95% CI=1.05–3.56; P=0.034), and GSE103479 (HR=2.37; 95% CI=1.34–4.21; P=0.003) datasets (Table III). Furthermore, data stratification analysis on GSE24549 patients indicated that the module was independent of stage (P=1.50×10−2 for stage II group; P=1.51×10−2 for stage III group; Fig. 8A and B), as well as the GSE24551 patients (P=1.30×10−2 for stage II group; P= 6.97×10−2 for stage III group; Fig. 8C and D) and GSE103479 patients (P=1.76×10−2 for stage II group; P=1.37×10−2 for stage III group; Fig. 8E and F). In addition, in terms of OS survival in GSE103479 patients, the module was independent of the patient age and the mutation of P53 (P=4.84×10−2 for patients aged >65 years; P=1.52×10−2 for the P53 mutated group; Fig. 8G and H). Based on these data, it is concluded that the 14-inflammatory-gene-module is a strong prognostic indicator for CRC.
Table III.
Univariate and multivariate Cox regression analysis of the module gene signature and disease-free survival of colorectal cancer patients in the training and test Gene Expression Omnibus datasets.
| Variables | Univariate model
|
Multivariate model
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI of HR | P-value | HR | 95% CI of HR | P-value | |
| GSE24550 (DFS) | ||||||
| Module risk score | 2.17 | 1.42–3.31 | <0.001 | 2.13 | 1.36–3.33 | 0.001 |
| Stage | 1.77 | 0.72–4.35 | 0.216 | 1.12 | 0.43–2.90 | 0.817 |
| GSE24549 (DFS) | ||||||
| Module risk score | 1.28 | 0.99–1.66 | 0.052 | 1.24 | 0.97–1.57 | 0.080 |
| Stage | 2.02 | 1.11–3.71 | 0.022 | 1.93 | 1.05–3.56 | 0.034 |
| GSE24551 (DFS) | ||||||
| Module risk score | 1.88 | 1.42–2.47 | <0.001 | 1.76 | 1.34–2.32 | <0.001 |
| Stage | 1.97 | 1.19–3.25 | 0.008 | 1.61 | 0.96–2.70 | 0.069 |
| GSE103479 (OS) | ||||||
| Module risk score | 2.13 | 1.24–3.64 | 0.006 | 1.58 | 0.92–2.70 | 0.094 |
| Stage | 2.18 | 1.30–3.67 | 0.003 | 2.37 | 1.34–4.21 | 0.003 |
| Age | 1.06 | 1.03–1.09 | <0.001 | 1.05 | 1.02–1.09 | 0.001 |
| Gender | 0.97 | 0.59–1.62 | 0.919 | 1.26 | 0.71–2.26 | 0.428 |
| BRAF mutation | 1.91 | 0.90–4.07 | 0.091 | 1.17 | 0.50–2.78 | 0.714 |
| KRAS mutation | 0.87 | 0.51–1.51 | 0.629 | 0.97 | 0.53–1.77 | 0.913 |
| P53 mutation | 0.57 | 0.33–0.98 | 0.041 | 0.68 | 0.39–1.19 | 0.173 |
HR, hazard ratio; CI, confidence interval; DFS, disease-free survival; OS, overall survival.
Figure 8.
Stratification analysis of independent datasets based on the stage. Kaplan-Meier survival curves of patients stratified by stage based on the 14-inflammatory-gene-module are shown for patients in the three datasets. (A) Stage II (n=46) and (B) stage III (n=37) CRC patients DFSin GSE24549; (C) stage II (n=90) and (D) stage III (n=70) CRC patients DFS in GSE24551; (E) stage II (n=83) and (F) stage III (n=72) CRC patients OS in GSE103479. In addition, (G) Kaplan-Meier OS curves of patients (n=102) in the GSE103479 dataset stratified by age (>65 years) based on the 14-inflammatory-gene-module are displayed. (H) Kaplan-Meier OS curves of patients (n=78) in the GSE103479 dataset stratified by P53 mutation based on the 14-inflammatory-gene-module are presented. DFS, disease-free survival; OS, overall survival.
Discussion
Inflammatory genes serve a key role in cell-cell communication, and dysfunctional inflammatory response can cause various diseases, including cancer. Thus, deciphering the inflammation-mediated mechanisms in CRC can provide opportunities for the early detection and treatment of CRC. A variety of inflammation-associated genes in CRC have been previously studied. For instance, Hickish et al (45) revealed that MABp1 was associated with an antitumor activity and relief of debilitating symptoms in patients with advanced CRC. Foersch et al (46) reported that inhibition of VEGFR2 signaling led to senescence of human CRC, which may improve the CRC patient survival in clinical practice. Zhu et al (47) also suggested that STING mediated protection against colorectal tumorigenesis by participating in intestinal inflammation.
In the present study, a systematical analysis on the inflammatory genes in CRC was performed, developing an integrated network-based strategy to construct and identify the inflammation-associated networks and modules. The current study built two networks, namely IRNN and IRDN, with inflammatory genes in the IRNN closely interacting with cancer genes, and genes in the IRDN enriched in numerous cancer-associated functions and pathways. These inflammatory genes may modulate the communication between cancer and normal cells. In addition, the current study highlighted the importance of inflammatory genes in the prognosis of CRC through the identification of a 14-inflammatory-gene-module, which was further validated by three independent datasets, providing a novel insight for CRC diagnosis and therapy.
Notably, the top six hub nodes in IRNN were all inflammatory genes, including EP300, which encodes the E1A-binding protein p300 and is important in the processes of cell proliferation and differentiation. The frame-shift mutations of EP300 have been reported to contribute to cancer pathogenesis by deregulating E1A-binding protein p300-mediated functions (48). Furthermore, the tyrosine kinases of the SRC family participate in oncogenic signaling in advanced CRC, which is crucial in the progression of CRC (49). The CBP protein (encoded by CREBBP), together with p300, may influence colonic cell physiology by affecting Wnt signaling, as well as affect colonic tumorigenesis and stem cell pluripotency (50). In addition, Ding et al (51) observed that Grb2-associated binder 2 (Gab2) promoted intestinal tumor metastasis by inducing epithelial-to-mesenchymal transition through the MEK/ERK/MMP pathway, which indicated that Gab2 may function as a novel prognostic factor for CRC patients. Additionally, with the immunohistochemistry study, researchers have found that MAPK is highly expressed and extremely active in colorectal cells and can be a therapeutically relevant target in inflammatory bowel disease (52,53).
In conclusion, although the inflammation-associated genes and modules should be confirmed by further experimental validations, the present study indeed provided a new direction for the exploration of the genesis and development of CRC using two inflammation-mediated gene networks. Furthermore, a 14-inflammatory-gene-module with prognostic potential was identified.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus, Gene Ontology and the Human Protein Reference Database repositories, www.ncbi.nlm.nih.gov/geo, geneontology.org and www.hprd.org, respectively.
Authors' contributions
ZS conceived and designed the experiments. HJ, LD, FG, YG, HZ and DF performed data analysis. ZS and HJ wrote this manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Kishiki T, Lapin B, Matsuoka H, Watanabe T, Takayasu K, Kojima K, Sugihara K, Masaki T. Optimal surveillance protocols after curative resection in patients with stage IV colorectal cancer: A multicenter retrospective study. Dis Colon Rectum. 2018;61:51–57. doi: 10.1097/DCR.0000000000000950. [DOI] [PubMed] [Google Scholar]
- 3.van der Stok EP, Spaander MCW, Grünhagen DJ, Verhoef C, Kuipers EJ. Surveillance after curative treatment for colorectal cancer. Nat Rev Clin Oncol. 2017;14:297–315. doi: 10.1038/nrclinonc.2016.199. [DOI] [PubMed] [Google Scholar]
- 4.Kahi CJ, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, Lieberman D, Levin TR, Robertson DJ, Rex DK, United States Multi-Society Task Force on Colorectal Cancer Colonoscopy surveillance after colorectal cancer resection: Recommendations of the US Multi-society task force on colorectal cancer. Gastroenterology. 2016;150:758–768.e11. doi: 10.1053/j.gastro.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Cooper JA, Parsons N, Stinton C, Mathews C, Smith S, Halloran SP, Moss S, Taylor-Phillips S. Risk-adjusted colorectal cancer screening using the FIT and routine screening data: Development of a risk prediction model. Br J Cancer. 2018;118:285–293. doi: 10.1038/bjc.2017.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westwood M, Lang S, Armstrong N, van Turenhout S, Cubiella J, Stirk L, Ramos IC, Luyendijk M, Zaim R, Kleijnen J, Fraser CG. Faecal immunochemical tests (FIT) can help to rule out colorectal cancer in patients presenting in primary care with lower abdominal symptoms: A systematic review conducted to inform new NICE DG30 diagnostic guidance. BMC Med. 2017;15:189. doi: 10.1186/s12916-017-0944-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passamonti B, Malaspina M, Fraser CG, Tintori B, Carlani A, D'Angelo V, Galeazzi P, Di Dato E, Mariotti L, Bulletti S, et al. A comparative effectiveness trial of two faecal immunochemical tests for haemoglobin (FIT). Assessment of test performance and adherence in a single round of a population-based screening programme for colorectal cancer. Gut. 2018;67:485–496. doi: 10.1136/gutjnl-2016-312716. [DOI] [PubMed] [Google Scholar]
- 8.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 9.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 10.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 11.van Geel RM, Beijnen JH, Bernards R, Schellens JH. Treatment individualization in colorectal cancer. Curr Colorectal Cancer Rep. 2015;11:335–344. doi: 10.1007/s11888-015-0288-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakurai T, Kashida H, Watanabe T, Hagiwara S, Mizushima T, Iijima H, Nishida N, Higashitsuji H, Fujita J, Kudo M. Stress response protein cirp links inflammation and tumorigenesis in colitis-associated cancer. Cancer Res. 2014;74:6119–6128. doi: 10.1158/0008-5472.CAN-14-0471. [DOI] [PubMed] [Google Scholar]
- 15.Waldner MJ, Neurath MF. Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin Immunol. 2014;26:75–79. doi: 10.1016/j.smim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Cao AT, Cong Y. Microbiota regulation of inflammatory bowel disease and colorectal cancer. Semin Cancer Biol. 2013;23:543–552. doi: 10.1016/j.semcancer.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho YA, Lee J, Oh JH, Chang HJ, Sohn DK, Shin A, Kim J. Genetic variation in PARGC1A may affect the role of diet-associated inflammation in colorectal carcinogenesis. Oncotarget. 2017;8:8550–8558. doi: 10.18632/oncotarget.14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Zhou Z, Zhang X, Zheng L, He D, Ye Y, Zhang QQ, Qi CL, He XD, Yu C, et al. Inflammatory molecule, PSGL-1, deficiency activates macrophages to promote colorectal cancer growth through NFκB signaling. Mol Cancer Res. 2017;15:467–477. doi: 10.1158/1541-7786.MCR-16-0309. [DOI] [PubMed] [Google Scholar]
- 19.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Lenferink AE, Deng Y, Collins C, Cui Q, Purisima EO, O'Connor-McCourt MD, Wang E. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun. 2010;1:34. doi: 10.1038/ncomms1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sveen A, Agesen TH, Nesbakken A, Rognum TO, Lothe RA, Skotheim RI. Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Med. 2011;3:32. doi: 10.1186/gm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sveen A, Ågesen TH, Nesbakken A, Meling GI, Rognum TO, Liestøl K, Skotheim RI, Lothe RA. ColoGuidePro: A prognostic 7-gene expression signature for stage III colorectal cancer patients. Clin Cancer Res. 2012;18:6001–6010. doi: 10.1158/1078-0432.CCR-11-3302. [DOI] [PubMed] [Google Scholar]
- 23.Agesen TH, Sveen A, Merok MA, Lind GE, Nesbakken A, Skotheim RI, Lothe RA. ColoGuideEx: A robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61:1560–1567. doi: 10.1136/gutjnl-2011-301179. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plaisier CL, Pan M, Baliga NS. A miRNA-regulatory network explains how dysregulated miRNAs perturb oncogenic processes across diverse cancers. Genome Res. 2012;22:2302–2314. doi: 10.1101/gr.133991.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub. Web Presence Working Group AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, Jiang W, Li W, Lian B, Chen X, Hua L, Lin H, Li D, Li X, Liu Z. Topological properties of the drug targets regulated by microRNA in human protein-protein interaction network. J Drug Target. 2011;19:354–364. doi: 10.3109/1061186X.2010.504261. [DOI] [PubMed] [Google Scholar]
- 32.Adamcsek B, Palla G, Farkas IJ, Derényi I, Vicsek T. CFinder: Locating cliques and overlapping modules in biological networks. Bioinformatics. 2006;22:1021–1023. doi: 10.1093/bioinformatics/btl039. [DOI] [PubMed] [Google Scholar]
- 33.Palla G, Derényi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alizadeh AA, Gentles AJ, Alencar AJ, Liu CL, Kohrt HE, Houot R, Goldstein MJ, Zhao S, Natkunam Y, Advani RH, et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood. 2011;118:1350–1358. doi: 10.1182/blood-2011-03-345272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, Li Y, Lu J, Pan T, Ding N, Wang Z, Shao T, Zhang J, Wang L, Li X. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 2015;43:8169–8182. doi: 10.1093/nar/gkv853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Attoub S, Rivat C, Rodrigues S, Van Bocxlaer S, Bedin M, Bruyneel E, Louvet C, Kornprobst M, André T, Mareel M, et al. The c-kit tyrosine kinase inhibitor STI571 for colorectal cancer therapy. Cancer Res. 2002;62:4879–4883. [PubMed] [Google Scholar]
- 40.Montero JC, Seoane S, Ocaña A, Pandiella A. Inhibition of SRC family kinases and receptor tyrosine kinases by dasatinib: Possible combinations in solid tumors. Clin Cancer Res. 2011;17:5546–5552. doi: 10.1158/1078-0432.CCR-10-2616. [DOI] [PubMed] [Google Scholar]
- 41.Blaj C, Schmidt EM, Lamprecht S, Hermeking H, Jung A, Kirchner T, Horst D. Oncogenic effects of high MAPK activity in colorectal cancer mark progenitor cells and persist irrespective of RAS mutations. Cancer Res. 2017;77:1763–1774. doi: 10.1158/0008-5472.CAN-16-2821. [DOI] [PubMed] [Google Scholar]
- 42.Barry GS, Cheang MC, Chang HL, Kennecke HF. Genomic markers of panitumumab resistance including ERBB2/HER2 in a phase II study of KRAS wild-type (wt) metastatic colorectal cancer (mCRC) Oncotarget. 2016;7:18953–18964. doi: 10.18632/oncotarget.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang SW, Hu J, Guo QH, Zhao Y, Cheng JJ, Zhang DS, Fei Q, Li J, Sun YM. AZD1480, a JAK inhibitor, inhibits cell growth and survival of colorectal cancer via modulating the JAK2/STAT3 signaling pathway. Oncol Rep. 2014;32:1991–1998. doi: 10.3892/or.2014.3477. [DOI] [PubMed] [Google Scholar]
- 44.Xiong H, Du W, Zhang YJ, Hong J, Su WY, Tang JT, Wang YC, Lu R, Fang JY. Trichostatin A, a histone deacetylase inhibitor, suppresses JAK2/STAT3 signaling via inducing the promoter-associated histone acetylation of SOCS1 and SOCS3 in human colorectal cancer cells. Mol Carcinog. 2012;51:174–184. doi: 10.1002/mc.20777. [DOI] [PubMed] [Google Scholar]
- 45.Hickish T, Andre T, Wyrwicz L, Saunders M, Sarosiek T, Kocsis J, Nemecek R, Rogowski W, Lesniewski-Kmak K, Petruzelka L, et al. MABp1 as a novel antibody treatment for advanced colorectal cancer: A randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2017;18:192–201. doi: 10.1016/S1470-2045(17)30006-2. [DOI] [PubMed] [Google Scholar]
- 46.Foersch S, Sperka T, Lindner C, Taut A, Rudolph KL, Breier G, Boxberger F, Rau TT, Hartmann A, Stürzl M, et al. VEGFR2 signaling prevents colorectal cancer cell senescence to promote tumorigenesis in mice with colitis. Gastroenterology. 2015;149:177–189.e10. doi: 10.1053/j.gastro.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Q, Man SM, Gurung P, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J Immunol. 2014;193:4779–4782. doi: 10.4049/jimmunol.1402051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim MS, Lee SH, Yoo NJ, Lee SH. Frameshift mutations of tumor suppressor gene EP300 in gastric and colorectal cancers with high microsatellite instability. Hum Pathol. 2013;44:2064–2070. doi: 10.1016/j.humpath.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 49.Sirvent A, Benistant C, Roche S. Oncogenic signaling by tyrosine kinases of the SRC family in advanced colorectal cancer. Am J Cancer Res. 2012;2:357–371. [PMC free article] [PubMed] [Google Scholar]
- 50.Bordonaro M, Lazarova DL. CREB-binding protein, p300 butyrate, and Wnt signaling in colorectal cancer. World J Gastroenterol. 2015;21:8238–8248. doi: 10.3748/wjg.v21.i27.8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding C, Luo J, Yu W, Gao S, Yang L, Chen C, Feng J. Gab2 is a novel prognostic factor for colorectal cancer patients. Int J Clin Exp Pathol. 2015;8:2779–2786. [PMC free article] [PubMed] [Google Scholar]
- 52.Lowenberg M, Verhaar A, van den Blink B, Ten Kate F, van Deventer S, Peppelenbosch M, Hommes D. Specific inhibition of c-Raf activity by semapimod induces clinical remission in severe Crohn's disease. J Immunol. 2005;175:2293–2300. doi: 10.4049/jimmunol.175.4.2293. [DOI] [PubMed] [Google Scholar]
- 53.Hardwick JC, van den Brink GR, Offerhaus GJ, van Deventer SJ, Peppelenbosch MP. NF-kappaB, p38 MAPK and JNK are highly expressed and active in the stroma of human colonic adenomatous polyps. Oncogene. 2001;20:819–827. doi: 10.1038/sj.onc.1204162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus, Gene Ontology and the Human Protein Reference Database repositories, www.ncbi.nlm.nih.gov/geo, geneontology.org and www.hprd.org, respectively.