Figure 1.
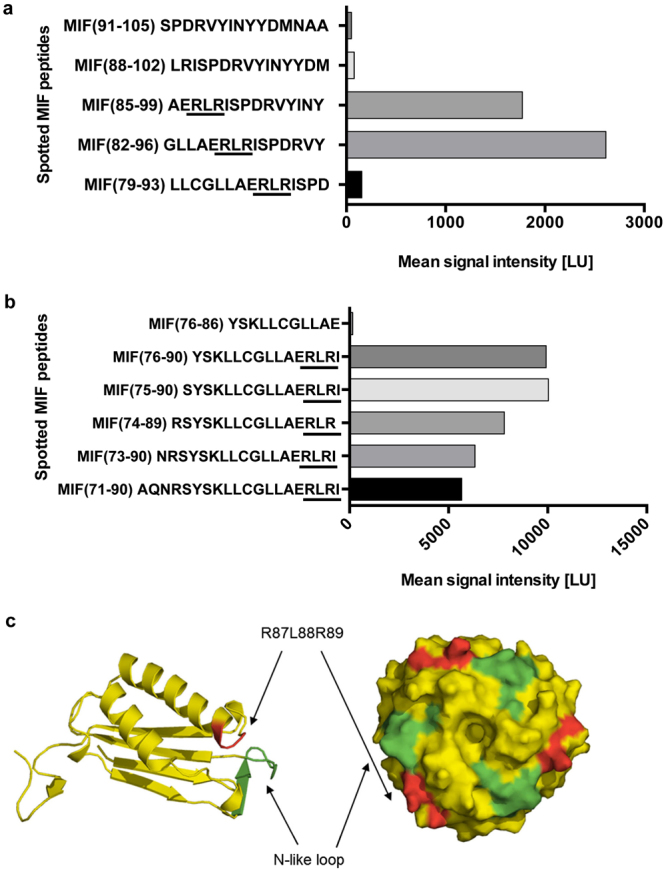
Identification of the RLR sequence as a potential MIF binding region to the N-terminal peptide of CXCR4. (a,b) The peptide spot microarray method suggests that the RLR tripeptide at sequence position 87–89 may contribute to MIF/CXCR4 binding. A peptide spot array containing 15-mer spotted MIF peptides positionally shifted by three amino acids were probed with biotin-CXCR4(1–27). Graphs are plots of spotted MIF peptides over the intensity of the binding signal to biotin-CXCR4(1–27) as read-out by streptavidin Cy5.5 fluorescence. (a) Of five positionally shifted 15-mer peptides of the region 79–105 only peptides containing RLR interact with CXCR4(1–27). (b) Binding of RLR-containing MIF peptides is modulated by N-terminal extension, but residues N-terminal of RLR do not exhibit binding activity per se. (c) Structural model of MIF (as monomer and trimer) and position of the N-like loop (green) and the RLR sequence (red). Note: in the three-dimensional conformation of the monomer, RLR is located in the vicinity of the N-like loop of MIF. The trimeric structure shows that both the N-like loop and RLR are surface-exposed on the trimer (see also Fig. 7).
