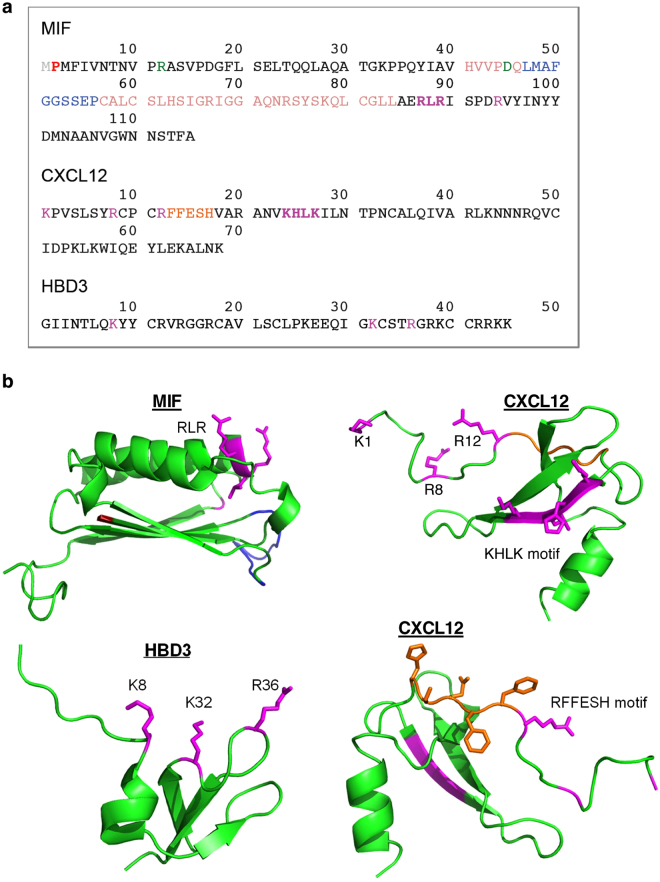
Figure 7.
Structure comparison of MIF, CXCL12, and human β-defensin-3. (a) Comparison of the amino acid sequences, highlighting residues/motifs that have been implicated in binding to CXCR4. MIF: RLR (magenta, bold), Arg-94 (R94; magenta), N-like loop (suggested to contribute to site 1 binding35; blue), Pro-2 (suggested to contribute to site 1 binding35; red), extended N-like loop (suggested to contribute to site 1 binding35; beige), pseudo-ELR motif (discontinuous, contributes to MIF/CXCR2 interface; green), Met-1 (cleaved upon expression; grey). CXCL12: KHLK motif (magenta, bold), Lys-1/Arg-8/Arg-12 charge cluster (magenta), RFFESH motif (orange). HBD3: Lys-8/Lys-32/Arg-36 charge cluster (magenta). (b) Comparison of the three-dimensional structures. Ribbon structures of the respective monomers are shown, with the backbones depicted in green. The corresponding color code and the captions for the residues/motifs that have been implicated in CXCR4 binding are as in (a); in the MIF structure, the ‘extension’ of the N-like loop (beige color in Fig. 7a) is not colored and the processed N-terminal methionine is not shown for clarity reasons. CXCL12 is depicted in two views (i) to highlight the KHLK motif and the charge cluster (top right) and (ii) the RFFESH motif (bottom right). Protein structures were produced/visualized with PyMOL.