Figure 7.
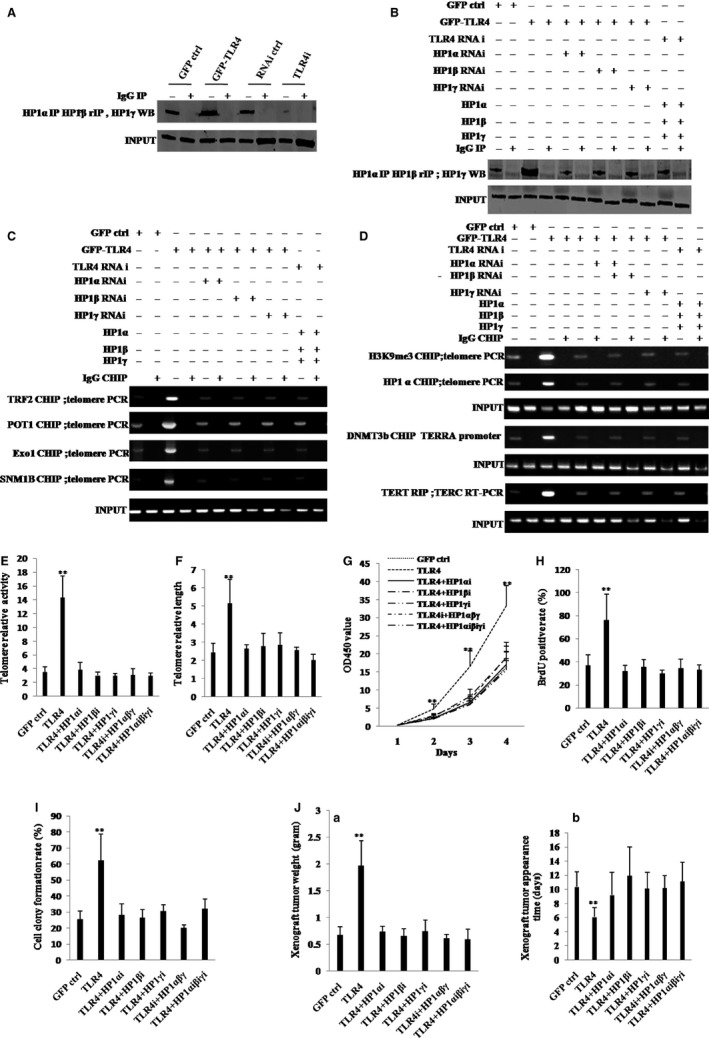
HP1 isoforms are required for TLR4 oncogenic action. A Repeat Co‐IP with anti‐HP1α or anti‐HP1β followed by Western blotting with anti‐HP1γ. IgG IP served as negative control. Western blotting with anti‐HP1γ served as INPUT. B, Repeat Co‐IP with anti‐HP1α or anti‐HP1β followed by Western blotting with anti‐HP1γ. IgG IP served as negative control. Western blotting with anti‐HP1γ served as INPUT. C, ChIP assay with anti‐TRF2, anti‐POT1, anti‐Exo1, anti‐SNM1B followed by PCR with telomere promoter primers. IgG ChIP served as negative control. PCR with telomere promoter primers served as INPUT. D, (upper) ChIP assay with anti‐H3K9me3, anti‐HP1α, anti‐DNMT3b followed by PCR with telomere or TERRA promoter primers. IgG ChIP served as negative control. PCR with telomere or TERRA promoter primers served as INPUT. (lower) RIP with anti‐TERT followed by RT‐PCR with TERC primers. IgG RIP served as negative control. RT‐PCR for TERC served as INPUT. B‐D, hLCSC lines: GFP ctrl group, TLR4 group, TLR4i group, and hLCSC lines whose HP1α, HP1β and HP1γ are overexpressed or depleted. The even number lanes are results from IgG controls. E, Telomerase activity assay with TRAP method primers. F, Real‐time PCR detection of telomere length. G, Cell growth assay using CCK8. H, Cell BrdU staining assay. I, Soft‐agar colony formation assay. J, (a) The wet weight of xenografted tumours from mouse. (b) PCNA staining (DAB staining, original magnification × 100). E‐J, hLCSC lines: GFP ctrl group, TLR4 group, TLR4 group transfected with pGFP‐V‐RS‐HP1α, pGFP‐V‐RS‐HP1β, pGFP‐V‐RS‐HP1γ and pGFP‐V‐RS‐HP1(α, β, γ), and TLR4i group transfected with pcDNA‐(HP1α, HP1β and HP1γ). Each value was presented as mean ± standard error of the mean (SEM). Mean ± SEM. **P < .01; *P < .05. For all Western blotting, we repeated the experiments for three times. We measured grey value of the bands for quantification. Each value was presented as mean ± standard error of the mean (SEM) (Student's t test)
