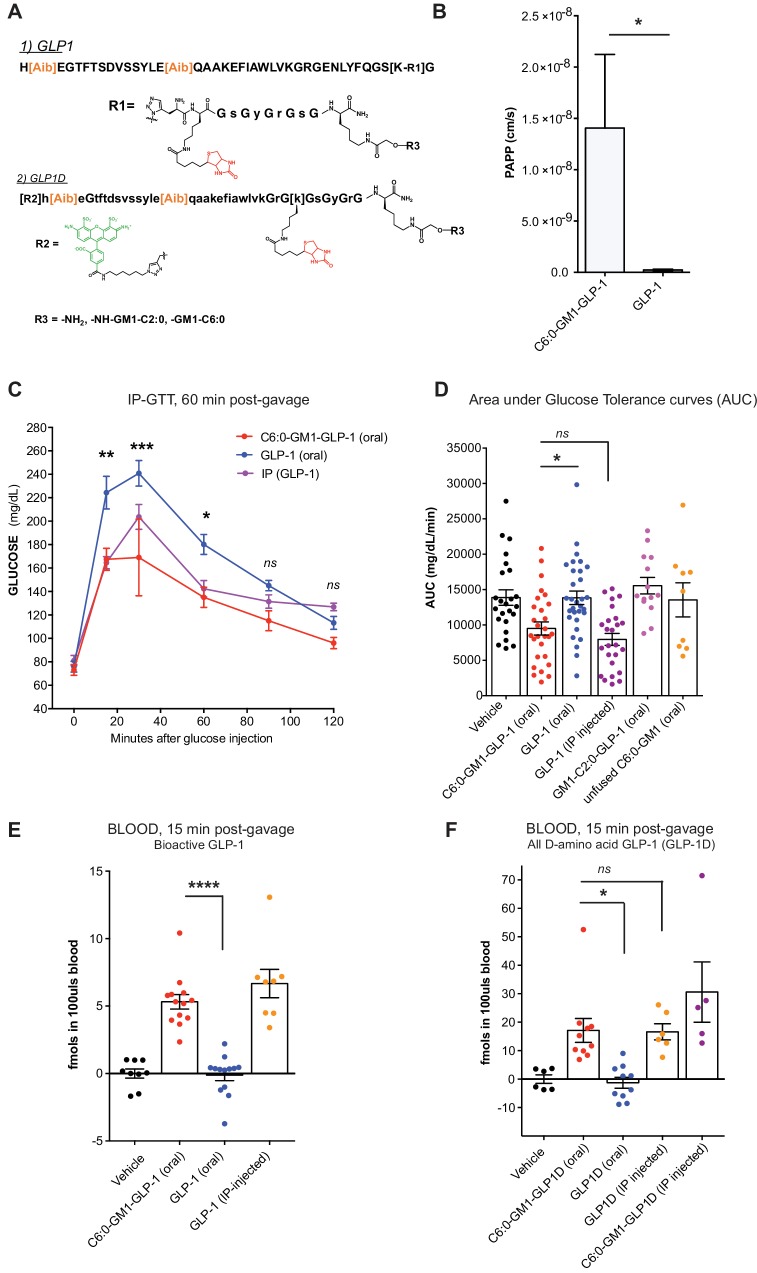
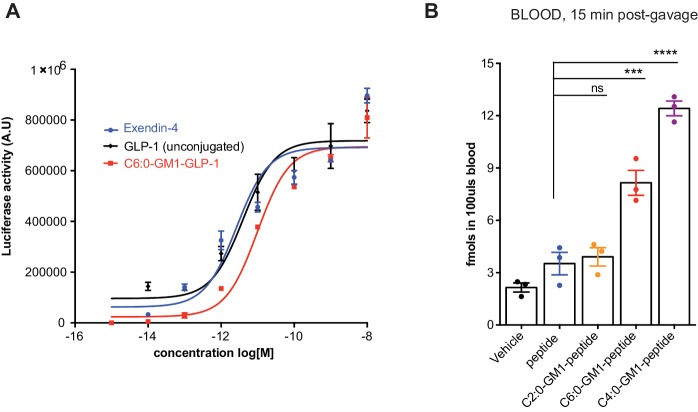
Figure 5. GM1-mediated absorption of GLP-1 affects blood glucose metabolism.
(A) GLP-1 and all-D GLP-1 isomer sequence used for coupling to a C6:0 and C2:0 GM1 ceramide species. (B) In vitro transcytosis assay with C6:0-GM1-GLP-1, or unfused GLP-1 across T84 cell monolayers (three independent experiments) (Unpaired t-test, *p<0.5). (C) Representative intraperitoneal glucose tolerance test after gastric gavage of 10 nmol/kg C6:0-GM1-GLP-1. Each point represents mean ± s.e.m (n = 4 mice). Mice fed C6:0-GM1-GLP-1 (red curve) show faster recovery after a glucose challenge in contrast to mice gavaged with unfused GLP-1 (blue curve) or vehicle (black curve) (D) Effect of the indicated GM1-GLP-1 species, GLP-1 alone, or vehicle on glucose tolerance quantified as Area under Curve (AUC) for 10 independent experiments with each data point representing individual mice and bar representing the mean ± s.e.m. (E) GLP-1 in blood 15 min after gastric gavage quantified for each species using the luciferase bioassay (fmols of compound per 100uls blood for three independent experiments). (F) Systemic absorption of an all D-isomer of GLP-1 used to directly measure the cargo in blood (three independent experiments). (A–E) Each data point on graphs represents individual mice and bars represent mean ± s.e.m. (ns = non significant, *p<0.5; ***p<0.0001, Tukey’s multiple comparison test).