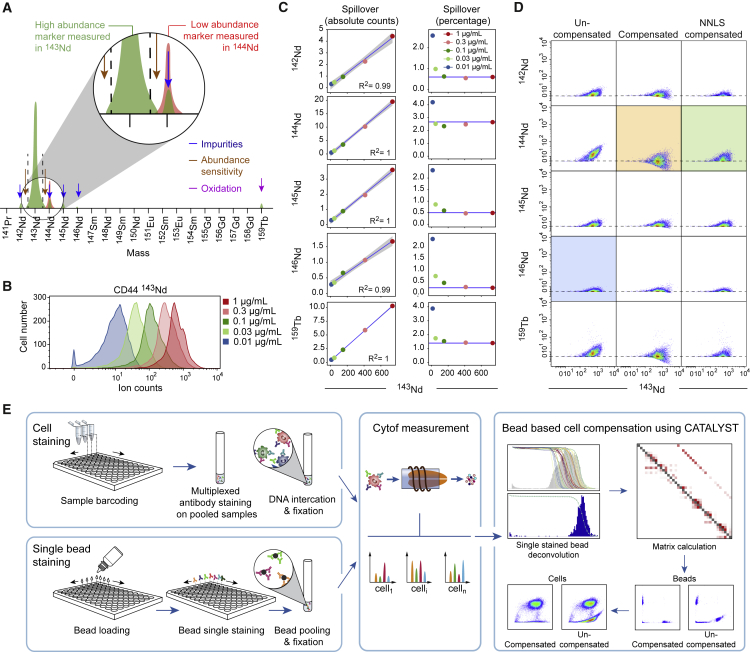
Figure 1.
Properties of Mass Cytometry Spillover and Description of a Single-Stained Bead-Based Compensation Workflow
(A) Schematic view of sources of signal interference that affect mass cytometry.
(B) Histogram showing signal intensity upon staining of PBMCs with the indicated concentrations of anti-CD44 antibody.
(C) The panels on the left display, as scatterplots, the median intensities of signals obtained in the main channel (143Nd) and spillover-affected channels for PBMCs stained with anti-CD44 antibody. Linear models are shown as blue lines. For each relationship, the coefficient of determination is indicated. The panels on the right display the spillover percentage calculated for each concentration. The blue lines indicate the spillover as assessed based on the highest antibody concentration.
(D) Scatterplots showing the signals of the anti-CD44 antibody in the main channel and in the spillover-affected channels before compensation (left column), after compensation with the conventional fluorescent flow cytometry approach (middle column), and after compensation with the NNLS method (right column). The green box shows how the NNLS compensation better preserves the data structure of a channel unaffected by spillover (blue box) than standard flow cytometry compensation does (orange box).
(E) Depiction of the workflow used to correct for spillover. Staining of control antibody-capture beads and samples are performed in parallel. Single-stained beads are pooled, and mass cytometry data are acquired on the beads and the samples. The CATALYST R package enables identification of the single-positive bead populations, calculates the compensation matrix, and applies the matrix to correction of sample data for spillover.