Figure 4.
Spillover Affects IMC Data and Can Be Corrected Using Our Compensation Strategy
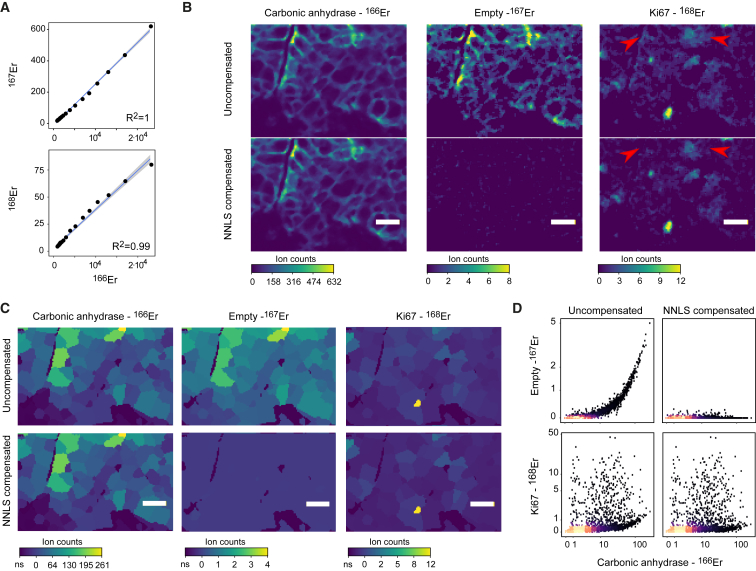
(A) Binning the signals of an imaged 166Er metal spot (to the 95th percentile of the 166Er pixel values) into 20 bins with equal pixel numbers shows a linear relationship between 166Er and 167Er over several orders of magnitude (upper panel). The relationship between 166Er and 168Er appears linear but saturates at the higher counts (lower panel).
(B) Representative image section of a breast cancer tissue sample imaged by IMC. Top row shows uncompensated images of 166Er (used to label antibody to carbonic anhydrase), 167Er (no antibody labeled with this metal), and 168Er (used to label anti-KI67). The bottom row shows corresponding NNLS compensated images. For visualization, a 3 × 3 pixel median filter was used to reduce noise. Scale bars, 20 μm. Red arrows indicate part of the image where low signal was removed by compensation.
(C) Segmentation mask shown on representative images described in (B). The mean pixel intensities of the signals observed in the indicated channels per cell are displayed.
(D) Scatterplots from single-cell segmentation data from the IMC images before (left) and after (right) compensation. Arcsinh transformed ion counts (cofactor of 2) are shown.