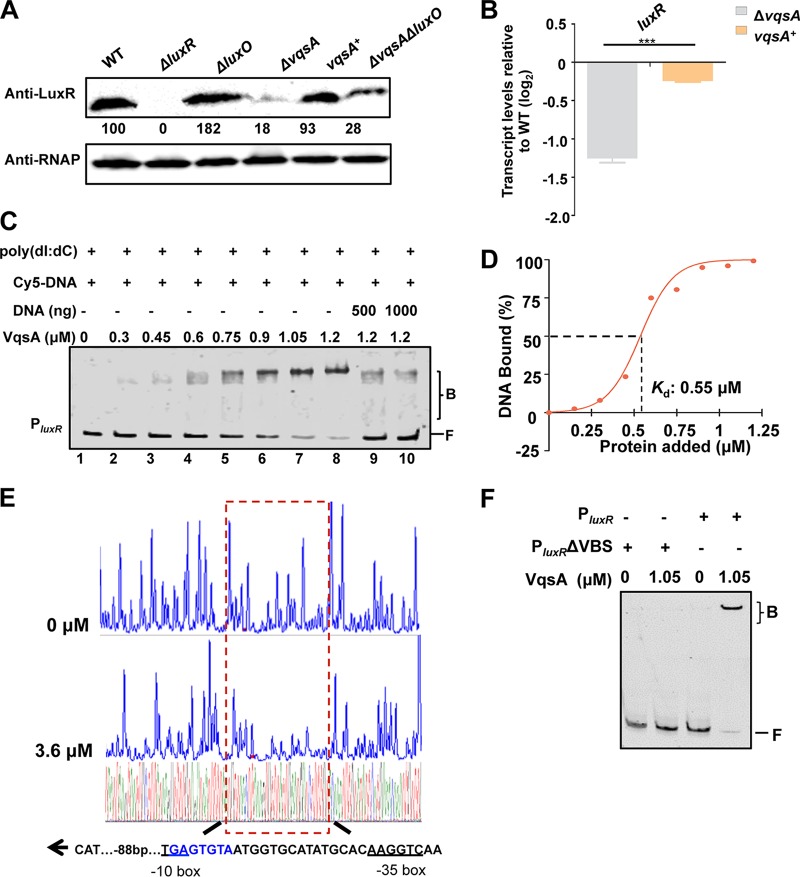
FIG 2.
VqsA regulates luxR expression by directly binding to the luxR promoter. (A) Western blot analysis showing LuxR expression in WT and ΔluxR, ΔluxO, ΔvqsA, vqsA+, and ΔvqsA ΔluxO mutant cells grown in LBS for 9 h. RNAP was used as a loading control. The numbers indicate the densitometric measurements. (B) qRT-PCR analysis of luxR transcripts in the WT, ΔvqsA mutant, and vqsA+ mutant cells grown in LBS for 9 h; 16S rRNA was selected as a control. The results are displayed as the mean ± SD (n = 3). ***, P < 0.001 based on Student's t test. (C) EMSA examination of the binding of VqsA to PluxR. The amount of VqsA protein used was as indicated, and 20 ng of each Cy5-labeled probe was added to the EMSA reactions. The shifts were verified to be specific in experiments in which 25- to 50-fold excess of unlabeled specific DNA and nonspecific competitor DNA [poly(dI:dC)] were used. (D) Plot showing the binding affinity of VqsA for the luxR promoter. The densitometric intensities of bound DNA fragments were plotted against VqsA concentrations. The indicated dashed line represents the concentration of VqsA that caused half-maximal binding (Kd). (E) DNase I footprinting analysis of VqsA binding to a binding site in the luxR promoter (shown in the dashed box). Electropherograms show a DNase I digestion of the PluxR promoter probe after incubation with 0 or 3.6 μM VqsA. The corresponding nucleotide sequences that were protected by VqsA are indicated below, with the binding motif highlighted in blue. The direction of transcription (as depicted with an arrow), the location of the partial −10 box and −35 box (underlined), and the VqsA-binding site relative to the start codon ATG are shown. (F) EMSA investigation of the binding of VqsA to the specific VqsA-binding site (VBS) in the luxR promoter. The PluxR variant lacking the VBS sequence (PluxR ΔVBS) was also investigated.