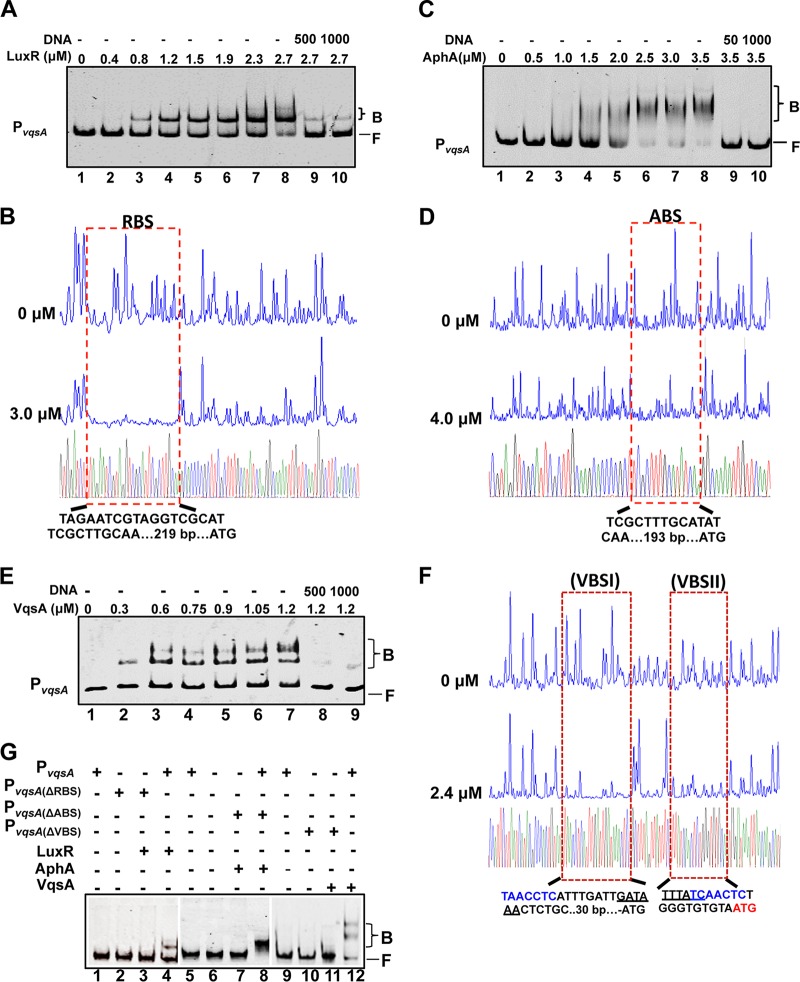
FIG 4.
AphA, LuxR, and VqsA bind to PvqsA. (A, C, and E) EMSAs were performed with purified LuxR (A), AphA (C), and VqsA (E). The amounts of the proteins used are indicated, and 20 ng of each Cy5-labeled PvqsA probe was added to the EMSA reactions. The shifts were verified to be specific in experiments in which 25- to 50-fold excess of unlabeled specific DNA and nonspecific competitor DNA [poly(dI:dC)] were added. (B, D, and F) DNase I footprinting analysis of LuxR (B), AphA (D), or VqsA (F) binding to their distinct binding sites in PvqsA. Electropherograms of a DNase I digestion of 400 ng of the PvqsA promoter probe after incubation with 0 or 3.0 μM LuxR, 0 or 4.0 μM AphA, and 0 or 2.4 μM VqsA are shown. The nucleotide sequences that were protected by LuxR, AphA, or VqsA are as indicated below. The specific palindromic sequences are underlined. Dashed boxes indicate the binding site revealed by DNase I footprinting assays. (G) EMSA investigation of the binding of LuxR, AphA, or VqsA to their specific binding sites, RBS, ABS, and VBS, in the luxR promoter region. The WT and the variant PvqsA lacking the related binding sites were also investigated.