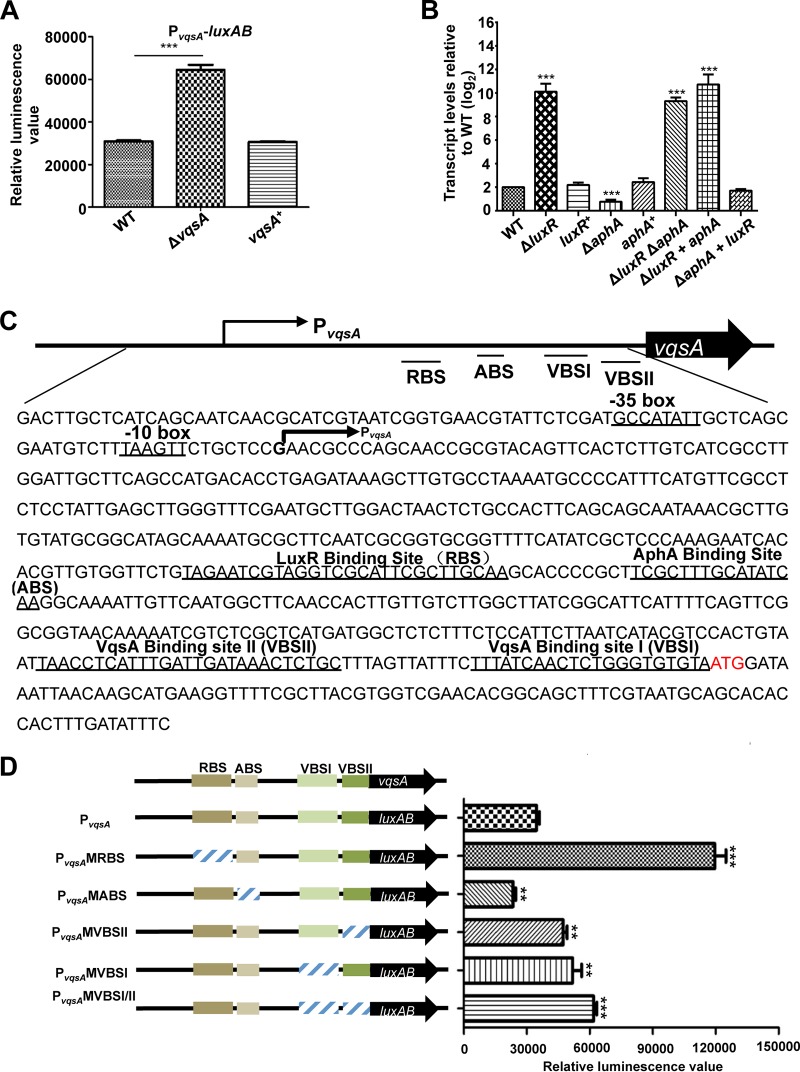
FIG 5.
The expression of vqsA is regulated by AphA, LuxR, and VqsA at the transcriptional level. (A) Transcriptional analysis of PvqsA-luxAB in the WT and ΔvqsA mutant strains cultured in LBS for 9 h. The results are shown as the mean ± SD (n = 3). ***, P < 0.001 based on Student's t test. (B) qRT-PCR analysis of the vqsA transcripts in ΔluxR, ΔaphA, luxR+, and aphA+ mutants and in the ΔluxR ΔaphA double mutant, as well as the related strains exhibiting epistatic expression (ΔluxR aphA mutant and ΔaphA luxR mutant) cultured in LBS for 9 h, and mRNA transcripts were detected by qRT-PCR; 16S rRNA was selected as a control. The results are displayed as the mean ± SD (n = 3); **, P < 0.01; ***, P < 0.001, based on Student's t test relative to the WT results. (C) The nucleotide sequence of PvqsA. The LuxR-binding site (RBS), AphA-binding site (ABS), VqsA-binding site I (VBSI), and VqsA-binding site II (VBSII) and the −10 box and −35 box are underlined. The PvqsA transcription start site as determined by 5′-RACE is labeled with an arrow, and the translational start codon (ATG) is in red. (D) The promoter activities of PvqsA and its variants fused to luxAB. Fluorescence levels were assayed for each of the strains. The data are shown as the mean ± standard error of the mean (SEM) (n = 3). **, P < 0.01; ***, P < 0.001, based on Student's t test relative to the WT PvqsA promoter.