ABSTRACT
Nitrite has been used as a bacteriostatic agent for centuries in food preservation. It is widely accepted that this biologically inert molecule functions indirectly, serving as a stable reservoir of bioactive nitric oxide (NO) and other reactive nitrogen species to impact physiology. As a result, to date, we know surprisingly little about in vivo targets of nitrite. Here, we carry out comparative analyses of nitrite and NO physiology in Escherichia coli and in Shewanella oneidensis, a Gram-negative environmental bacterium renowned for respiratory versatility. These two bacteria differ from each other in many aspects of nitrite and NO physiology, including NO generation, NO degradation, and unexpectedly, their contrary susceptibility to nitrite and NO. In cell extracts of both bacteria, most of the NO targets are also susceptible to nitrite, and vice versa. However, with respect to growth inhibition caused by NO, the targets are impacted distinctly; NO targets are responsible for the inhibition of growth of E. coli but not of S. oneidensis. More surprisingly, all proteins identified to be implicated in NO tolerance in other bacteria appear to play a dispensable role in protecting S. oneidensis against NO. These data suggest that S. oneidensis is equipped with a robust but yet unknown NO protecting system. In the case of nitrite, it is clear that the target of physiological significance in both bacteria is cytochrome heme-copper oxidase.
IMPORTANCE Nitrite is toxic to living organisms at high levels, but such antibacterial effects of nitrite are attributable to the formation of nitric oxide (NO), a highly reactive radical gas molecule. Here, we report that Shewanella oneidensis is highly resistant to NO but sensitive to nitrite compared to Escherichia coli by approximately 4-fold. In both bacteria, nitrite inhibits bacterial growth by targeting cytochrome heme-copper oxidase. In contrast, the targets of NO are diverse. Although these targets are similar in E. coli and S. oneidensis, they are responsible for growth inhibition caused by NO in the former but not in the latter. Overall, the presented data, along with the previous data, solidify a proposal that the in vivo targets of NO and nitrite in bacteria are largely different.
KEYWORDS: nitrite, nitric oxide, cytochrome c oxidase, stress response
INTRODUCTION
Nitrite is the central player in the nitrogen biogeochemical cycle by linking nitrate to gas nitrogen or ammonium. During the reduction to nitrogen, nitrite is converted to nitric oxide (NO), a molecule that has been intensively studied because of its diverse roles, particularly those that are beneficial, in the physiologies of bacteria and eukaryotes (1, 2). Both nitrite and NO can damage cells, causing nitrosative stress (3), but they differ from each other fundamentally in their chemistries (1). Unlike nitrite, NO by itself not only is a reactive free radical but also reacts with oxygen and superoxide to generate a family of reactive nitrogen species (RNS) under aerobic conditions (3). In addition, NO also differs from nitrite in that it can easily diffuse into cells. Because of these features, it has been widely accepted that the antibacterial effects of nitrite are attributable to nitric oxide (NO) formation (1, 4). There are two ways for bacterial cells to generate NO endogenously, an ability which is restricted to certain species at present (1, 2). One depends on bacterial NO synthases (bNOSs), which catalyze the conversion of l-arginine to l-citrulline with consumption of NADPH and O2. The other is through the process of respiratory denitrification by either copper-containing nitrite reductases or heme-containing cytochrome (cyt) cd1 nitrite reductases. To date, many targets of NO have been identified, including proteins with redox-active centers, lipids, and DNA (2, 5). In the gammaproteobacteria Escherichia coli and Salmonella enterica serovar Typhimurium, the proteins susceptible to NO include aconitase, dihydroxy-acid dehydratase, α-ketoglutarate dehydrogenase, fructose-1,6-biphosphate aldolase, argininosuccinate synthase, lipoamide dehydrogenase, and some components of the respiratory chain (6–12). Because of this, the ultimate phenotype caused by NO is growth inhibition (2).
To cope with NO, bacteria have evolved multiple strategies (2). The most efficient one is to directly remove the gas molecule. In bacteria, the enzymes known to decompose NO include flavohemoglobin (Hmp) functioning as nitric-oxide dioxygenase (NOD), nitric oxide reductase (NOR), flavorubredoxin (NorV), flavodiiron proteins (such as flavin reductase Fre of Escherichia coli), and the hybrid cluster protein (Hcp) (13–18). Apart from these, periplasmic c552 nitrite reductase (NrfA) can also function as a NO scavenger in certain bacteria, in addition to catalyzing the respiratory reduction of nitrite to ammonia (19, 20). Recently, it has been shown that several single domain globins (SDGs), often called “truncated” hemoglobins, have been implicated in NO metabolism, functioning as enzymatic NODs when coupled to suitable electron donors (21, 22). In addition to NO scavengers, another important strategy is to produce NO-resistant proteins that protect the cellular iron pool against NO stress (8, 12, 23). For example, Vibrio cholerae NnrS, an NO-regulated heme-containing protein, elevates NO resistance by protecting the cellular iron-pool and iron-sulfur enzymes from NO inhibition (23).
Shewanella oneidensis, a Gram-negative gammaproteobacterium, is renowned for its respiratory versatility, capable of respiring a variety of organic and inorganic substrates as electron acceptors (EAs), including nitrate and nitrite (24). This feature has been largely attributable to a large number of c-type cyts (up to 42), which are mainly involved in energy transduction processes as electron carriers (25–27). The c-type cyts are generally susceptible to nitrosative stress agents, such as nitrite and NO, which can bind iron and subsequently inhibit function (1, 28). Consistently, nitrite has been found to be extremely toxic to S. oneidensis cells grown under either aerobic or anaerobic conditions. While the primary target of nitrite for oxygen respiration is heme-copper oxidase (HCO) cyt cbb3, nitrite inhibition of alternative EA respiration involves cAMP-Crp (catabolite repression protein) regulation and NapB, a soluble c-type cyt that, in excess, dissipates electrons of the quinol pool (29–31).
In our previous studies, nitrite and NO were found to differ from each other in their influences on S. oneidensis growth, and more importantly, nitrite inhibition was not affected by the addition of an NO scavenger (30, 32). This prompted us to investigate the biology of nitrite and NO in bacteria. Here, we report that E. coli and S. oneidensis are distinct from each other in their susceptibilities to nitrite and NO: the former is more resistant to nitrite but more sensitive to NO than the latter by approximately 4-fold. Unlike E. coli, S. oneidensis cannot generate NO endogenously and lacks the most efficient NO scavengers. In both bacteria, the primary cellular target of nitrite is HCO, which dictates aerobic growth. Although the NO targets are common between E. coli and S. oneidensis, they are accountable for growth inhibition in the former but not in the latter. Overall, these data suggest that S. oneidensis possesses a robust system protecting it from protein damage caused by NO.
RESULTS
Distinct susceptibilities of S. oneidensis and E. coli to nitrite and NO.
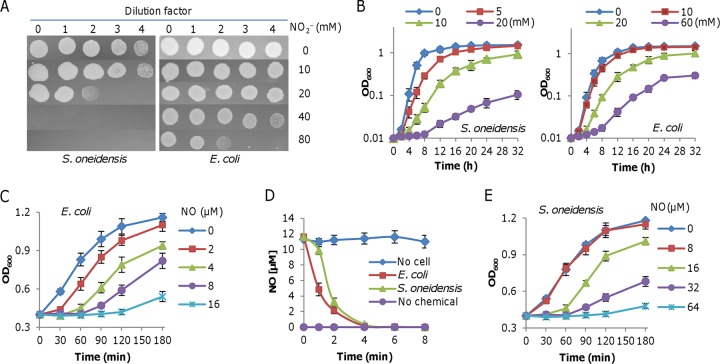
Previously, we demonstrated that S. oneidensis is highly susceptible to nitrite (NaNO2 and KNO2) during aerobiosis (29–32). For comparison, we assessed the susceptibility of E. coli to nitrite under the same conditions (Fig. 1A). On LB agar plates, E. coli was able to grow in the presence of 80 mM nitrite, whereas S. oneidensis failed to do so with 40 mM. Evidently, 80 mM nitrite was required to impair the growth of E. coli to a level comparable to that of S. oneidensis with 20 mM. In parallel, the inhibitory impacts of nitrite on growth in liquid media were investigated. Cultures at mid-log phase (optical density at 600 nm [OD600] of ∼0.4, the same throughout the study) were inoculated in fresh media containing nitrite of various levels to an OD600 of ∼0.01 and growth was monitored. Consistently, the inhibition effect of nitrite on S. oneidensis was much stronger than that on E. coli (Fig. 1B). These data indicate that E. coli is more resistant to nitrite than S. oneidensis.
FIG 1.
Nitrite and NO physiologies in S. oneidensis and E. coli. (A) Nitrite susceptibility assay. Cells of S. oneidensis and E. coli wild-type strains were grown in LB to the mid-log phase. Serial dilutions were prepared with fresh LB, and 5 μl of each dilution was dropped on plates containing nitrite at indicated concentrations. Results were photographed 24 h later. (B) Inhibition of nitrite at various concentrations on growth in liquid medium. (C) Impacts of NO released from DEA-NONOate on growth of the mid-log-phase E. coli cultures. DEA-NONOate was added to cultures adjusted to an OD600 of ∼0.4 to indicated concentrations and growth was monitored. (D) NO consumption assay. NO released from 16 μM DEA-NONOate was mixed with cells prepared as in panel C, and NO concentrations were monitored. (E) Impacts of NO released from DEA-NONOate on growth of the mid-log-phase S. oneidensis cultures performed as in panel C. Shown are either representative data or means ± SEMs from at least three experiments.
Given that nitrite and NO, as nitrosative stress agents, are proposed to exert their deleterious impacts on proteins by similar mechanisms (28) and, more importantly, that antibacterial effects of nitrite are attributable to NO formation (1, 4), we expected similar trends in NO susceptibility from these two bacteria. The impacts of NO on growth were assessed with sodium (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate ([DEA-NONOate] half-life [t1/2] ≈ 2 min) as reported before (6, 33). The growth of E. coli cells at the mid-log phase was significantly inhibited by DEA-NONOate at a concentration as low as 2 μM, and no growth was detected for 2 h with 16 μM (Fig. 1C). In an analysis of NO consumption, NO concentrations up to 12 μM were recorded when 16 μM DEA-NONOate was added to the cell-free medium under anaerobic conditions, whereas there was no detectable NO in the absence of the chemical (Fig. 1D). Consistent with a previous finding (6), NO was degraded rapidly by E. coli cells, such that it could only be detected within the first 6 min (Fig. 1D). Surprisingly, S. oneidensis cells that were similarly prepared showed much stronger tolerance to NO. The inhibitory effects became evident when 16 μM DEA-NONOate was present, and it required 64 μM to exert an impact comparable to that imposed by 16 μM on E. coli (Fig. 1E). This robust resistance is apparently not attributable to NO removal, because NO degradation was not faster in S. oneidensis than in E. coli (Fig. 1D).
Additionally, the inhibitory impacts of NO on growth in liquid media were estimated with 2,2′-(hydroxynitrosohydrazino)bis-ethanamine ([DETA-NONOate] t1/2 ≈ 20 h) used to deliver NO to the culture, because NO can be stably released during the measuring period (33). The experiments were performed as for the nitrite treatment. In media containing DETA-NONOate that were stabilized for 10 h, cultures at the mid-log phase were inoculated to an OD600 of ∼0.01 and the growth was monitored. We found that the inhibition effect of NO on growth of these two bacteria appeared similar to that of nitrite, but in S. oneidensis, it required four times more NO to match the growth inhibition observed in E. coli (see Fig. S1 in the supplemental material). Altogether, these data indicate that E. coli and S. oneidensis differ from each other significantly in their resistances to nitrite and NO.
S. oneidensis does not generate NO endogenously.
To address the contrasting susceptibilities of S. oneidensis to nitrite and NO, we attempted to characterize the related aspects in NO biology, especially NO generation and tolerance, in S. oneidensis. It has been reported that S. oneidensis is able to produce NO in the presence of nitrite under anaerobic but not aerobic conditions (34). If so, bNOSs could not be the enzyme for NO production, as they require oxygen as a necessary cosubstrate (35) and there are no homologues to bNOS in the genome (36). Moreover, homologues of cyt cd1 nitrite reductases, NO-producing enzymes in denitrifying bacteria, are not found in the genome either.
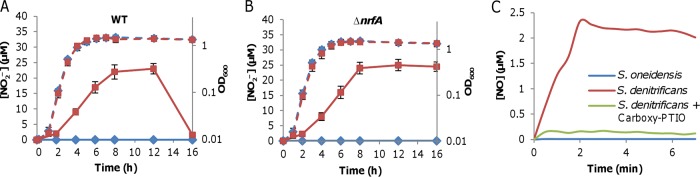
To search for new enzymatic sources of NO, we first made attempts to confirm the NO-forming ability of S. oneidensis. NO levels in cultures grown aerobically were estimated with the established approach that measures an oxidized product of NO, nitrite (37). In cultures grown in rich medium, nitrite levels were determined in the wild type and in the ΔnrfA mutant, which cannot convert nitrite to ammonium (38). Nitrite was not detected in cultures of both strains grown under aerobic conditions (Fig. 2A and B), indicating that NO is not formed under aerobic conditions. For confirmation, we expressed the nosA gene of Bacillus subtilis in S. oneidensis under the control of the S. oneidensis arcA promoter, which is constitutively active (39, 40). This time, nitrite was detected in both wild-type and ΔnrfA cultures grown under aerobic conditions (Fig. 2A and B). The concentrations of nitrite increased with the cell densities until the stationary phase, reached a maximum of ∼25 μM, and then started to decline in the wild type due to the reduction of nitrite to ammonium but remained the same in the ΔnrfA strain, a phenomenon reported previously (32, 41).
FIG 2.
S. oneidensis may not generate NO endogenously. (A and B) Growth and NO production of wild-type (WT) (A) and ΔnrfA (B) strains in LB under aerobic conditions. Growth and samples were measured and collected at indicated time points, and nitrite concentrations in supernatants were determined. Dashed and solid lines represent growth and nitrite concentrations, respectively. Samples of cells without and with the Bacillus subtilis nosA gene are in blue and red, respectively. (C) Representative NO production of S. oneidensis and S. denitrificans under anaerobic conditions. Cultures of all strains grown on TMAO to an OD600 of ∼0.3 were collected, washed, and suspended in the same medium containing 5 mM nitrite. Data for NO production from both strains in the presence of NO scavenger carboxy-PTIO at 0.1 mM were included. The dissolved NO concentrations were monitored using an NO-specific electrode. Shown are either representative data or means ± SEMs from at least three experiments.
To determine NO generation under anaerobic conditions, mid-log-phase wild-type cultures (OD600 of ∼0.2) grown on trimethylamine N-oxide (TMAO) were collected by centrifugation, washed, and resuspended to the same OD600 values in fresh medium containing 5 mM NO2−, and NO production was determined by using an NO-specific electrode. As shown in Fig. 2C, NO concentrations were not above the detection limit (∼50 nM). To confirm this, we performed the same experiment with Shewanella denitrificans strain OS217, a verified denitrifying bacterium (42). NO reduced from NO2− by this organism was detected, up to 2.4 μM. The production of NO was further verified with the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide, potassium salt (carboxy-PTIO), which quenched the NO signal to below measurable levels. Altogether, these data show that S. oneidensis does not generate NO endogenously.
NO tolerance systems in S. oneidensis.
S. oneidensis is able to consume NO, on the basis of the significant difference in NO concentrations between growing cultures and cell-free control medium (43). However, the genome does not encode flavohemoglobin or flavorubredoxin for NO removal; the potential candidates remaining for the role include cyt c nitrite reductase NrfA, the hybrid cluster protein Hcp (SO_1363), single domain globin SO_0039, flavodiiron protein Fre (SO_0504), and NO protecting protein NnrS (SO_2805) (38, 44–47).
To determine the roles of these proteins in NO tolerance (degradation and protection), we first determined whether their expression is responsive to NO. By using an integrated lacZ reporter, the activities of the promoters for these five genes under various culture conditions were assessed (Fig. 3A). In line with previous data (30, 32), we found that the nrfA gene in mid-log-phase cells grown on oxygen or TMAO was not active compared to that in cells grown on both TMAO and nitrite. While SO_0039 in oxygen-respiring cells was expressed nearly 3 times higher than in those respiring on TMAO, the fre gene displayed a contrasting pattern: low and increased expression under aerobic and anaerobic conditions, respectively, implying that these two genes are likely expressed only under specific conditions. Neither nrfA, fre, nor SO_0039 was responsive to NO, but nrfA was induced by nitrite, suggesting that these three genes may not be involved in NO biology. In the case of hcp and nnrS, we observed similar expression patterns, which were distinct from those of the other three. The expression levels of both hcp and nnrS in mid-log-phase cells increased drastically in cultures supplemented with DETA-NONOate. A similar upregulation in expression for these two genes was also observed from cultures containing nitrite. Given that both hcp and nnrS genes are responsive to NO as well as nitrite, they probably play a significant role in the cellular response to nitrosative stress. It is worth mentioning that there was a modest increase in the expression of both hcp and nnrS genes when cells grew in medium free of NO or nitrite under anaerobic conditions. The expression of these genes was also estimated by using real-time reverse transcription-quantitative PCR (qRT-PCR), and similar results were obtained (see Fig. S2).
FIG 3.
Impacts of NrfA, SO_0039, Hcp, and NnrS on NO resistance and removal. (A) Promoter activity measurement of PnrfA, PSO0039, Phcp, PnnrS, and Pfre by an integrated lacZ reporter in cells grown under indicated conditions. Cells of mid-log-phase cultures were pelleted, processed, and subjected to a β-galactosidase activity assay. O2, aerobic growth fumarate; +NO, containing 400 μM DETA-NONOate; TMAO, 30 mM; +NO2−, 5 mM nitrite. (B) Effects of NrfA on NO-induced growth inhibition. Growth of WT and ΔnrfA strains in LB containing 0.4 mM DETA-NONOate was compared. Expression of nrfA (ΔnrfA/pnrfA) was driven by Ptac promoter with 1 mM IPTG, resulting in at least 15× induction compared to the chromosomal copy in the WT as calibrated previously. (C) Effects of NrfA on NO consumption. Data are shown as means± SEMs from at least three experiments.
We then constructed in-frame deletion mutants for hcp, SO_0039, and nnrS and characterized the resulting mutants along with the ΔnrfA mutant with respect to growth and NO consumption. All mutants were indistinguishable from the wild type; for simplicity, the results of only the ΔnrfA mutant are presented and discussed (Fig. 3B), whereas representatives of other mutants are given in Fig. S3A. In the absence and presence of 400 μM DETA-NONOate, the growth of the ΔnrfA mutant was comparable to that of the wild type, indicating that the mutation does not impact the growth supported by oxygen respiration regardless of NO. Despite this, it was clear that NO significantly reduced the growth rates for both strains. Additionally, we found that there was no difference in NO consumption (Fig. 3C). For confirmation, we forced nrfA expression by using IPTG (isopropyl-â-d-thiogalactopyranoside)-inducible promoter Ptac to examine the effects of NrfA on NO (48). The expression system is effective, validated previously by Western blotting (49, 50). However, the forced expression of the nrfA gene by up to 1 mM IPTG, which resulted in more than 15-fold overexpression, had no effect on either growth or NO consumption of the ΔnrfA strain (Fig. 3C). Thus, we conclude that NrfA is dispensable in the degradation of or the protection from NO in S. oneidensis.
To check that the mutation of a single gene could be compensated by enhanced expression of the others, we monitored the expression of all these genes in the absence of one of the others (Fig. S3B). The results revealed that the expression of all of these genes was hardly affected by the individual loss of the other genes. Moreover, we constructed hcp and nnrS double mutants because their expression is responsive to NO. Similar to each single mutant, the double mutant was not distinguishable from the wild type under all test conditions, with respect to both growth and NO consumption (Fig. S3C). On the basis of all of these data, we conclude that none of these proteins plays an important role in NO tolerance in S. oneidensis.
HCOs are primary targets of nitrite but not of NO in S. oneidensis.
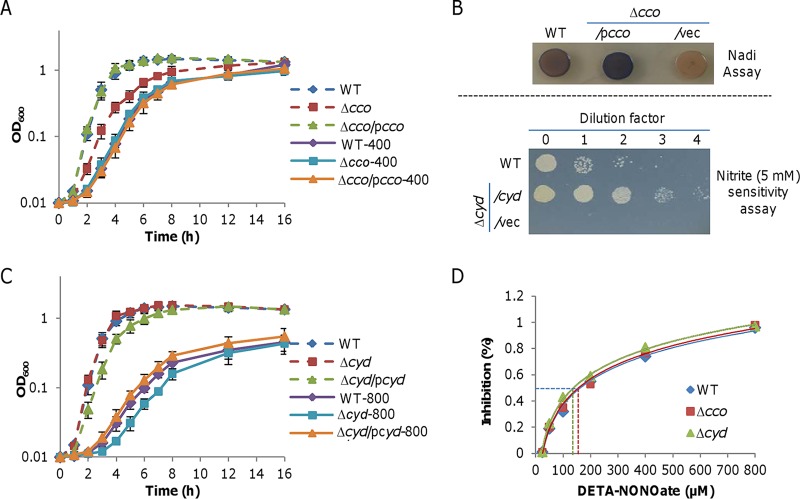
Previously, we have demonstrated that S. oneidensis cbb3 HCO is hypersensitive to nitrite (29). Given that HCOs are highly susceptible to NO in model bacteria E. coli and S. Typhimurium (8, 12), we reasoned that S. oneidensis cbb3 HCO is likely also sensitive to NO. The loss of cbb3 HCO, slowing the growth in the absence of NO as revealed before (51), did not worsen growth inhibition by NO released from up to 400 μM DETA-NONOate (Fig. 4A). Given that cyt bd is the only oxidase remaining functioning (51), the data suggest that cyt bd is sufficient to carry out oxygen respiration under NO stress. We then examined the ability of cbb3 HCO when overproduced to alleviate the NO inhibition. The overproduction of cbb3 HCO from IPTG-inducible promoter Ptac corrected the growth defect of the cbb3 HCO-deficient (Δcco) strain (Fig. 3A) and, more importantly, increased the oxidase activity substantially (Fig. 4B, upper panel), revealed by the Nadi assay that specifically detects cyt c oxidase-dependent respiration (52). Nevertheless, the growth inhibition by NO was not affected much by excessive cbb3 HCO (Fig. 4A).
FIG 4.
Oxidases are not important in S. oneidensis NO physiology. (A) Impact of cytochrome cbb3 HCO (encoded by the cco operon) on NO-induced growth inhibition. Growth of WT and Δcco strains in LB without or with 400 μM DETA-NONOate was compared. Cytochrome cbb3 HCO production (Δcco/pcco) was driven by Ptac with IPTG up to 1 mM (the data with 1 mM are shown). Data are shown as means ± SEMs from at least three experiments. (B) Representative data of overproduced cytochrome cbb3 HCO and bd (encoded by the cyd operon), presented in upper and lower panels, respectively. Upper panel: effects of overproduced cytochrome cbb3 HCO on cytochrome c oxidase activities revealed by the Nadi assay. Cells of indicated strains were grown in LB to the mid-log phase, and 5 μl of each culture was spotted on LB plates containing 1 mM IPTG and incubated for 16 h at 30°C. The Nadi assay was then performed and results were photographed 2 min after the reaction began. WT and previously verified Δcco strains served as positive and negative controls. Lower panel: nitrite sensitivity assay. Cells of indicated strains were grown in LB to the mid-log phase. Serial dilutions were prepared with fresh LB, and 5 μl of each dilution was dropped on plates containing 5 mM nitrite and 1 mM IPTG. Results were photographed 24 h later. (C) Impacts of cytochrome bd on NO-induced growth inhibition. Growth of WT and Δcyd strains in LB without or with 800 μM DETA-NONOate was compared (no difference was observed with 400 μM DETA-NONOate). Cytochrome bd production (Δcyd/pcyd) was performed as in panel A. (D) Representative IC50s of cytochrome cbb3 HCO and bd for DETA-NONOate. Respiration rates of membranes were measured in the presence of DETA-NONOate of increasing concentrations. Shown are either representative data or means ± SEMs from at least three experiments.
The growth of the bd-deficient (Δcyd) strain was only slightly affected by NO released from 800 μM but not from 400 μM DETA-NONOate, in sharp contrast to its hypersensitivity to nitrite (Fig. 4B and C). When overproduced, cyt bd modestly inhibited the growth due to the low efficacy of the enzyme (53) but substantially enhanced nitrite resistance (Fig. 4C). On the contrary, the growth of S. oneidensis in the presence of 800 μM DETA-NONOate was hardly affected by excessive cyt bd. These observations indicate that cyt bd confers S. oneidensis resistance to nitrite but not to NO, implying that NO may not specifically inhibit cbb3 HCO.
To further address whether cbb3 HCO is more sensitive than cyt bd to NO, we determined their half-maximal inhibitory concentrations (IC50s) for DETA-NONOate as for nitrite (50). Membrane preparations of S. oneidensis strains expressing only one of the terminal oxidases were used to measure oxygen reduction with ubiquinol-1 for cyt bd or with a combination of ascorbate and N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (TMPD) for cbb3 HCO (8). At ∼70 μM O2, the IC50 of S. oneidensis cyt bd was approximately 163 μM, whereas the IC50 for cbb3 HCO was approximately 140 μM (Fig. 4D). No difference in the IC50s between the wild-type and the Δcco strains was observed. As it has been soundly established that HCOs are hypersensitive to NO (8, 12), we hence hypothesize that the similar IC50s for cbb3 HCO and cyt bd are probably due to an unknown primary protection system that shields the enzymes from NO.
Growth-critical targets of nitrite and NO are likely different in E. coli and S. oneidensis.
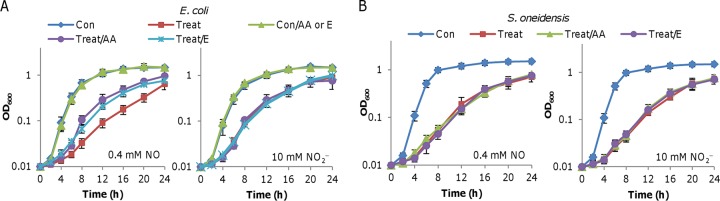
As reported before and above, HCOs are susceptible to nitrite and NO (8, 12, 29, 50). We have previously shown that HCOs are predominantly responsible for the growth inhibition by nitrite in both E. coli and S. oneidensis (29). However, the inhibitory effects of NO on growth are apparently not by the inactivation of these enzymes. In E. coli, dihydroxy-acid dehydratase (IlvD), an Fe-S cluster enzyme essential for branched-chain amino acid (BCAA) biosynthesis, has been identified as a critical target of NO by using an integrated network analysis; as a consequence, NO exposure induces transient BCAA auxotrophy (6). To test whether the protein is also a nitrite target, we examined the effects of BCAA addition on growth. The supplementation of all BCAAs partially corrected the growth inhibition caused by NO, but did not show any suppression of nitrite inhibition (Fig. 5A). Additionally, we found that an increased expression of E. coli IlvD (EcIlvD) induced by 1 mM IPTG, validated by an approximately 9-fold increase in enzyme activity (see Fig. S4A), elevated the resistance to NO but not to nitrite (Fig. 5A). These data suggest that EcIlvD is unlikely a critical target of nitrite under test conditions.
FIG 5.
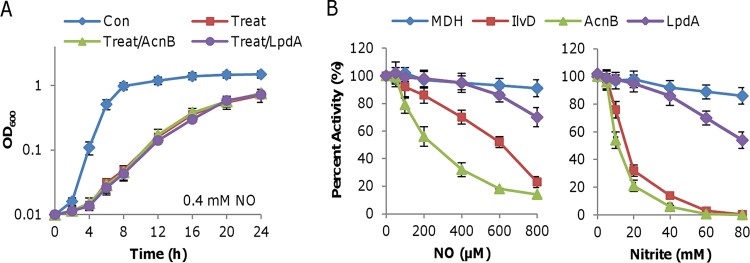
Targets of nitrite and NO in S. oneidensis and E. coli. Effects of BCAA addition or overproduction of IlvD on growth of E. coli (A) and S. oneidensis (B). Con, grown in LB; Treat, grown in LB with either 0.4 mM NO or 10 mM nitrite; Treat/AA, LB containing a mixture of BCAA; Treat/E, cells with overproduced respective IlvD in the presence of 1 mM IPTG. Data are shown as means ± SEMs from at least three experiments.
In the case of S. oneidensis, neither BCAA addition nor S. oneidensis IlvD ([SoIlvD] BLASTp E value against EcIlvD, 0.0) overproduction showed a significant impact on the growth of cultures in the presence of nitrite (Fig. 5B). Interestingly, these treatments were not effective in lessening the inhibition by NO (Fig. 5B). Moreover, we overproduced EcIlvD in S. oneidensis, but it did not elicit any significant difference with respect to growth (Fig. S4B). These data imply that IlvD may not be a target of NO or nitrite in S. oneidensis.
Established NO targets of other gammaproteobacteria are not accountable for NO susceptibility in S. oneidensis.
We then examined the roles of two other enzymes in nitrite inhibition, aconitase (AcnB) and lipoamide dehydrogenase (LpdA), which have been established as NO targets in gammaproteobacteria (6, 10, 54). The impacts of overproduced enzymes on the growth inhibition caused by NO and nitrite in S. oneidensis were assayed. After their induction by 1 mM IPTG, which resulted in a ≥4-fold increase in enzyme activities (Fig. S4A), neither protein was able to elicit any noticeable difference in the growth of the wild type in the absence or presence of DETA-NONOate (Fig. 6A). Similar results were obtained in the presence of nitrite. Thus, these proteins may not be inhibited by nitrite or NO to the extent of physiological significance in S. oneidensis.
FIG 6.
Targets of nitrite and NO in S. oneidensis and E. coli. (A) Effects of overproduction of SoAcnB and SoLpdA on growth of S. oneidensis. (B) Effects of nitrite and NO on activity of putative targets in S. oneidensis. Enzyme activities in cell extracts with increasing concentrations of NO, released by DEA-NONOate, or nitrite were determined. Percent activity is normalized to unexposed cell extracts. Data are shown as means ± SEMs from at least three experiments.
Unexpectedly, none of the established NO targets in other gammaproteobacteria appear to be implicated in NO inhibition. As stated above, we hypothesized that there would be a protection system against NO in S. oneidensis. Thus, we would expect that NO could compromise the activities of these enzymes in cell extracts. Indeed, all these enzymes were subject to NO inhibition when their activities were directly assessed in cell extracts (Fig. 6B). Compared to malate dehydrogenase (MDH), an NO-resistant enzyme used as the control (10), increased concentrations of NO had modest effects on the activities of SoLpdA. In contrast, SoIlvD and SoAcnB lost their activities significantly more rapidly upon NO treatment. Interestingly, similar results were obtained from nitrite treatment (Fig. 6B). These data indicate that SoIlvD and SoAcnB are susceptible to both NO and nitrite, but SoLpdA is probably unaffected by these nitrosative agents.
DISCUSSION
In this study, we presented evidence to show that S. oneidensis is highly resistant to NO but sensitive to nitrite compared to E. coli by approximately 4-fold. Subsequent investigations revealed that the cellular targets of nitrite and NO, which underlie growth inhibition, are largely different between these two bacteria. Previously, we demonstrated that cyt bd confers S. oneidensis nitrite resistance, because the primary target of nitrite for aerobic growth is HCO cbb3 (29, 51). Coincidently, cyt bd in E. coli, whose HCO is cyt bo, plays a similar role in nitrite resistance (29). Hence, HCOs appear to be primary bacterial targets of nitrite during aerobiosis. This may not be surprising, because HCOs catalyze the same reaction and share similar basic principles underlying redox-driven proton translocation, although they are diverse in terms of subunit compositions, electron donors, and heme types (55). However, HCOs are not identified as NO targets critical to aerobic growth by integrated network analysis or other high-throughput profiling, although biochemical analyses have illustrated that they are vulnerable upon exposure to NO (6, 8, 10, 56).
Most of the growth-critical NO targets revealed before are essential redox centers, such as Fe-S clusters, heme, and protein thiols, exemplified by Fe-S-containing aconitase and dihydroxy-acid dehydratase and lipoamide dehydrogenase with a thiol active site (6, 10, 54). Despite this, NO targets differ significantly from one bacterium to another, even within phylogenetically closely related bacteria (10). This suggests that many unknown factors are involved in the interactions of NO with redox-active centers, which remain to be identified. Notably, most of these growth-critical NO targets are found to also be susceptible to nitrite in cell extracts, but their involvement in nitrite-induced growth inhibition is negligible. One explanation is that nitrite, unlike NO, could not diffuse across the inner membrane, which separates these cytoplasmic enzymes from nitrite.
Clearly, NO inhibits effectively most of S. oneidensis counterparts of the growth-critical NO targets identified in E. coli and S. Typhimurium (6, 54), such as AcnB and IlvD, when they are present in cell extracts. However, they are not the enzymes accountable for the NO-induced growth inhibition. Unlike E. coli, in which supplementation with BCAA completely reversed the NO-dependent growth inhibition, S. oneidensis is still subject to NO inhibition. This difference is attributable to either of two possibilities. One is that other targets are simultaneously affected by NO in S. oneidensis. The repertoire of NO targets has been expanding in recent years, owing to advances in high-throughput approaches, such as metabolomic profiling (23). It is certain that many more will be discovered in the future. The other possibility implicated by the data presented here is that there is a robust NO protection system, which conceivably confers a layer of protection against protein damage by NO.
Although little is known about this system, it apparently confers on S. oneidensis unusually high resistance to NO compared to that of E. coli (2). The S. oneidensis genome does not encode any of most efficient NO scavengers, such as Hmp, NorV, and NO-detoxifying hemoglobin (2, 43). Additionally, periplasmic nitrite reductase NrfA, which confers NO tolerance to a broader group of bacteria by directly reducing NO and hydroxylamine (17, 57–59), appears to lack such activity in S. oneidensis. On the basis of these data, we propose that the unknown system is able to scavenge NO.
In addition to NO scavenging, S. oneidensis utilizes the NO-responding regulator NsrR to coordinate the expression of genes involved in coping with NO stress (47). The predicted NsrR regulon of S. oneidensis is very small, containing only three operons, hcp-hcr, dnrN, and nnrS, all of which are conserved NO-responding members among many Gram-negative bacteria (2, 47). To date, there has been only one report about NnrS; V. cholerae NnrS seems to protect the cellular iron pool from the formation of dinitrosyl iron complexes without scavenging NO (23). In the case of Hcp, its significance in NO tolerance differs from species to species (2, 18, 60). Moreover, we found that the contributions of Hcp and NnrS to NO tolerance in S. oneidensis, if any, are rather limited. While more investigation is needed, it is possible that the processes involving these proteins vary from species to species in bacteria and that their roles in NO tolerance are overshadowed by the unknown protection system.
In this study, we provided multiple lines of evidence to show that S. oneidensis does not generate NO endogenously, a result contradicting the finding reported previously (34). First, S. oneidensis lacks the enzymatic sources of NO that have been solidly defined in bacteria. Second, NO generation in S. oneidensis was assayed under the same conditions as for the positive-control bacteria, which are equipped with either bNOS or denitrifying nitrite reductase. Third, the production of B. subtilis bNOS enabled S. oneidensis to generate NO. As the earlier result was obtained from cultures grown on nitrate or nitrite as the sole EA and the result in this study was from cell suspensions, the two findings could reflect culture differences (34). Thus, the formation of NO in the previous report may be attributed to the acidification of nitrite, a possibility also suggested by the authors.
MATERIALS AND METHODS
Bacterial strains, plasmids, PCR primers, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The information for the primers used for generating PCR products is given in Table 2. For genetic manipulation, E. coli and S. oneidensis strains were grown in lysogeny broth ([LB] Difco, Detroit, MI) at 37°C and 30°C, respectively. When needed, the growth medium was supplemented with chemicals at the following concentrations: 2,6-diaminopimelic acid (DAP), 0.3 mM; ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; and gentamicin, 15 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Host for cloning | Lab stock |
| E. coli MG1655 | Wild type | Coli Genetic Stock Center |
| E. coli WM3064 | Donor strain for conjugation, ΔdapA | W. Metcalf, UIUC |
| B. subtilis 168 | Source of bacterial bNOS | Bacillus Genetic Stock Center |
| S. denitrificans | Shewanella denitrifying strain OS217 | DSM 15013 |
| S. oneidensis | ||
| MR-1 | Wild type | ATCC 700550 |
| HG0039 | ΔSO_0039 derived from MR-1 | This study |
| HG0504 | Δfre derived from MR-1 | This study |
| HG1363 | Δhcp derived from MR-1 | This study |
| HGCCO | Δcco (ΔccoNOPQ) derived from MR-1 | 50 |
| HG2805 | ΔnnrS derived from MR-1 | This study |
| HG3980 | ΔnrfA derived from MR-1 | 38 |
| HGCYD | Δcyd (ΔcydABX) derived from MR-1 | 66 |
| Plasmids | ||
| pHGM01 | Apr, Gmr, Cmr, att-based suicide vector | 27 |
| pHG102 | Kmr, ParcA expression vector | 63 |
| pHGEI01 | Integrative E. coli lacZ reporter vector | 41 |
| pBBR-Cre | Helper vector for antibiotic marker removal | 29 |
| pHGE-Ptac | Kmr, IPTG-inducible Ptac expression vector | 48 |
| pHGE-Ptac-cco | Inducible expression of cco | 50 |
| pHGE-Ptac-cyd | Inducible expression of cyd | 50 |
| pHGE-Ptac-nrfA | Inducible expression of nrfA | 30 |
| pHGE-Ptac-acnB | Inducible expression of acnB | This study |
| pHGE-Ptac-ilvD | Inducible expression of ilvD | This study |
| pHGE-Ptac-lpdA | Inducible expression of lpdA | This study |
| pHG102-nosA | Forced expression of nosA | This study |
| pHGEI-PnrfA-lacZ | E. coli lacZ under the control of nrfA promoter | 30 |
| pHGEI-PSO0039-lacZ | E. coli lacZ under the control of SO_0039 promoter | This study |
| pHGEI-Phcp-lacZ | E. coli lacZ under the control of hcp promoter | This study |
| pHGEI-PnnrS-lacZ | E. coli lacZ under the control of nnrS promoter | This study |
| pHGEI-Pfre-lacZ | E. coli lacZ under the control of fre promoter | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
TABLE 2.
Primers used in this study
For physiological characterization, both LB and defined medium MS containing 0.02% (wt/vol) vitamin-free Casamino Acids and 30 mM lactate as the electron donor were used in this study, and consistent results were obtained (61). Fresh medium was inoculated with overnight cultures grown from a single colony by 1:100 dilution, and the growth was determined by recording the optical density of cultures at 600 nm (OD600). For anaerobic growth, mid-log-phase aerobic cultures were pelleted by centrifugation, purged with nitrogen, and suspended in fresh medium prepared anaerobically to an OD600 of ∼0.02. TMAO (20 mM), was used as the electron acceptor, because it supports good growth and respiration with it is immune to nitrite inhibition (30). To assay the effects of NO on growth, DETA-NONOate (t1/2, 20 h at 37°C and 56 h at 25°C) was used, because it releases NO slowly and can maintain a relatively steady NO concentration (34). For amino acid complementation, BCAAs (Val, Leu, and Ile) and M and K (Met and Lys, respectively) were supplemented at 0.3 mM.
Mutagenesis and complementation.
In-frame deletion strains were constructed according to the att-based fusion PCR method described previously (27). In brief, two fragments flanking the gene of interest were amplified with primers containing attB, gene-specific sequences, and complementary sequences and then were joined by a second round of PCR. The resulting fusion fragment was introduced into plasmid pHGM01 by site-specific recombination using BP Clonase (Invitrogen) and maintained in E. coli WM3064, which is a DAP auxotroph. The resulting mutagenesis vector, after being verified by sequencing, was transferred from strain WM3064 to relevant S. oneidensis strains by conjugation. The integration of the mutagenesis construct into the chromosome was selected for by gentamicin resistance and confirmed by PCR. Verified transconjugants were grown in LB in the absence of NaCl and plated on LB plates supplemented with 10% sucrose. Gentamicin-sensitive and sucrose-resistant colonies were screened by PCR for the deletion of the target gene. The mutants were then verified by sequencing the deletion region.
In-frame deletion strains from previous studies have been verified by genetic complementation (Table 1). The genetic complementation for S. oneidensis mutants newly constructed in this study that have a distinct phenotype was performed with pHGE-Ptac, which carries IPTG-inducible promoter Ptac (39, 48). The same systems were also used for heterogeneous complementation. The coding sequence of each gene under test was cloned by restriction enzyme digestion and ligation. After being verified by sequencing, the vectors were introduced into the relevant mutants for phenotypic assays.
Nitrite sensitivity assay.
Bacterial strains grown to the mid-log phase were adjusted to approximately 108 CFU/ml, followed by 10-fold serial dilutions. Ten microliters of each dilution was spotted onto LB plates containing nitrite. The plates were incubated at 30°C before being read. The assays were repeated at least three times.
Determination of NO2− and NO concentrations.
The concentrations of nitrite in culture supernatants were measured by a modified Griess assay (62) and by using ion chromatography ICS-5000 with IonPac AS19 (Thermo Scientific). To determine NO production in the presence of NO2−, the relevant strains were grown in LB to the mid-log phase (OD600 of ∼0.3), collected by centrifugation, washed, and resuspended in fresh LB to an OD600 of 0.3, which was followed by the addition of 5 mM (final concentration) NaNO2. NO production was monitored using an ISO-NOPMC Mark II electrode (WPI Instruments, Sarasota, FL) run through an MLT1122 analog adapter system (AD Instruments, Colorado Springs, CO) with standard curves generated according to the manufacturer's instructions.
Measurement of NO consumption.
The NO consumption assay was performed under anaerobic conditions, because autoxidation of NO at high concentrations is very fast in the presence of oxygen. To prepare NO-containing samples, 10 ml fresh LB made anaerobically was transferred into Hungate tubes of 150 mm with DEA-NONOate, sealed with a rubber stopper and a screw cap, and incubated at 30°C. Cells from the strains of interest were grown to the mid-log phase in LB (OD600 of ∼0.3), collected by centrifugation, washed, purged with nitrogen gas, and resuspended in fresh LB prepared anaerobically to an OD600 of ∼0.5. One milliliter of the cell suspension was injected into the stabilized NO-containing samples in Hungate tubes. The tubes were then placed in a 30°C water bath and shaken periodically while monitoring the NO concentrations with an ISO-NOPMC Mark II electrode, as described above.
Controlled expression of relevant genes.
To assess the effects of the genes of interest expressed at various levels on NO and nitrite physiologies, we amplified and placed each of them under the control of the constitutively active S. oneidensis arcA promoter within pHG102 or under the IPTG-inducible promoter Ptac within pHGE-Ptac (39, 40, 48, 63). The pHGE-Ptac expression system was calibrated previously (30, 64). PCR amplification was carried out with genomic DNAs from the S. oneidensis, E. coli, and B. subtilis wild-type strains as the templates with the primers listed in Table 2. The resulting PCR products were digested by restriction enzymes corresponding to the restriction enzyme sites included in the primers, were ligated to vectors with T4 DNA ligase, and were transformed into E. coli WM3064. After verification by sequencing, the vectors were transferred into the relevant strains via conjugation. Cells carrying the vectors of interest were grown in the media indicated in the text and/or figure legends in the presence of IPTG at various levels.
Expression analyses.
To estimate the expression of the genes of interest, a segment containing the target promoter was amplified from genomic DNA and inserted into pHGEI01 by restriction enzyme digestion and ligation (41). After being verified by sequencing, the resultant vector was transferred to relevant S. oneidensis strains by conjugation for integration into the chromosome. The antibiotic marker was then removed by using an established approach (29). Cells grown to the mid-log phase were collected, and β-galactosidase activity assays were performed with an assay kit as described previously (29).
The expression of the genes of interest was also assessed by using qRT-PCR analyses with an ΑΒΙ7300 96-well qRT-PCR system (Applied Biosystems) as described previously (65). The expression of each gene was determined from three replicates in a single real-time qRT-PCR experiment. The cycle threshold (CT) values for each gene of interest were averaged and normalized against the CT value of 16S rRNA, whose abundance was consistent from early exponential phase to stationary phase. The relative abundance (RA) of each gene compared to that of 16S rRNA was calculated using the equation RA = 2−ΔCT.
Oxidase activity assay.
Visual analysis of the cbb3 HCO activity was done by staining colonies with the agents for the Nadi assay. Nadi reactions were carried out by the addition of α-naphthol and N′,N′-dimethyl-p-phenylenediamine (DMPD) on LB agar plates (52). The colonies were timed for the formation of the indophenol blue.
Solubilized membranes were prepared for the quantitative analysis of oxidase activity as described previously (50). In brief, cell pellets were resuspended in 20 mM Tris-HCl (pH 7.6) supplemented with DNase I and protease inhibitors and disrupted by using a French press. After removing the debris and unbroken cells, the membranes were pelleted by ultracentrifugation for 1 h at 230,000 × g at 4°C and subsequently resuspended in 20 mM Tris-HCl (pH 7.6) with 5% glycerol to a protein concentration of 10 mg/ml. Solubilization was performed with n-dodecyl β-d-maltoside (DDM) to a final concentration of 1% (wt/vol) on a rotary tube mixer for 2 h at 4°C. The DDM-solubilized membranes were obtained by collecting the supernatant after ultracentrifugation for 1 h at 230,000 × g at 4°C. Oxidase activity was assayed as a measure of oxygen consumption rates using an OxyGraph oxygen electrode (Hansatech) using either ubiquinol-1 (1 mM) or N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride ([TMPD] 1 mM) as the electron donor at 35°C according to the methods described previously (8, 50). The IC50s of the cyt bd and cbb3 HCO for nitrite and NO were obtained from plots of the rates against their concentrations.
Bioinformatics and statistical analyses.
Homologues of proteins of interest were identified via a BLASTp search of the NCBI's nonredundant protein database, using the amino acid sequence as the query. Student's t tests were performed for pairwise comparisons. The values are presented as the means ± standard errors of the means (SEMs).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (41476105) and the Natural Science Foundation of Zhejiang Province (LZ17C010001).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00559-18.
REFERENCES
- 1.Maia LB, Moura JJG. 2014. How biology handles nitrite. Chem Rev 114:5273–5357. doi: 10.1021/cr400518y. [DOI] [PubMed] [Google Scholar]
- 2.Stern AM, Zhu J. 2014. An introduction to nitric oxide sensing and response in bacteria, p 187–220. In Sima S, Geoffrey MG (ed), Advances in applied microbiology. Academic Press, Cambridge, MA. [DOI] [PubMed] [Google Scholar]
- 3.Bowman LAH, McLean S, Poole RK, Fukuto JM. 2011. The diversity of microbial responses to nitric oxide and agents of nitrosative stress: close cousins but not identical twins, p 135–219. In Robert KP. (ed), Advances in microbial physiology. Academic Press, Cambridge, MA. [DOI] [PubMed] [Google Scholar]
- 4.Reddy D, Lancaster J, Cornforth D. 1983. Nitrite inhibition of Clostridium botulinum: electron spin resonance detection of iron-nitric oxide complexes. Science 221:769–770. doi: 10.1126/science.6308761. [DOI] [PubMed] [Google Scholar]
- 5.Vine CE, Cole JA. 2011. Unresolved sources, sinks, and pathways for the recovery of enteric bacteria from nitrosative stress. FEMS Microbiol Lett 325:99–107. doi: 10.1111/j.1574-6968.2011.02425.x. [DOI] [PubMed] [Google Scholar]
- 6.Hyduke DR, Jarboe LR, Tran LM, Chou KJY, Liao JC. 2007. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc Natl Acad Sci U S A 104:8484–8489. doi: 10.1073/pnas.0610888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Husain M, Bourret TJ, McCollister BD, Jones-Carson J, Laughlin J, Vázquez-Torres A. 2008. Nitric oxide evokes an adaptive response to oxidative stress by arresting respiration. J Biol Chem 283:7682–7689. doi: 10.1074/jbc.M708845200. [DOI] [PubMed] [Google Scholar]
- 8.Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, Cooper CE. 2009. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat Chem Biol 5:94–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- 9.Bourret TJ, Boylan JA, Lawrence KA, Gherardini FC. 2011. Nitrosative damage to free and zinc-bound cysteine thiols underlies nitric oxide toxicity in wild-type Borrelia burgdorferi. Mol Microbiol 81:259–273. doi: 10.1111/j.1365-2958.2011.07691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC. 2011. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar Typhimurium. Cell Host Microbe 10:33–43. doi: 10.1016/j.chom.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giuffrè A, Borisov VB, Mastronicola D, Sarti P, Forte E. 2012. Cytochrome bd oxidase and nitric oxide: from reaction mechanisms to bacterial physiology. FEBS Lett 586:622–629. doi: 10.1016/j.febslet.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Jones-Carson J, Husain M, Liu L, Orlicky DJ, Vázquez-Torres A. 2016. Cytochrome bd-dependent bioenergetics and antinitrosative defenses in Salmonella pathogenesis. mBio 7:e02052-. doi: 10.1128/mBio.02052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner AM, Martin LA, Gardner PR, Dou Y, Olson JS. 2000. Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin): the B10 tyrosine hydroxyl is essential for dioxygen binding and catalysis. J Biol Chem 275:12581–12589. doi: 10.1074/jbc.275.17.12581. [DOI] [PubMed] [Google Scholar]
- 14.Gardner AM, Helmick RA, Gardner PR. 2002. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J Biol Chem 277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- 15.Gardner PR. 2012. Hemoglobin: a nitric-oxide dioxygenase. Scientifica 2012:683729. doi: 10.6064/2012/683729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomes CM, Giuffrè A, Forte E, Vicente JB, Saraiva LM, Brunori M, Teixeira M. 2002. A novel type of nitric-oxide reductase: Escherichia coli flavorubredoxin. J Biol Chem 277:25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- 17.Mills PC, Rowley G, Spiro S, Hinton JCD, Richardson DJ. 2008. A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiology 154:1218–1228. doi: 10.1099/mic.0.2007/014290-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Vine CE, Balasiny BK, Rizk J, Bradley CL, Tinajero-Trejo M, Poole RK, Bergaust LL, Bakken LR, Cole JA. 2016. The roles of the hybrid cluster protein, Hcp and its reductase, Hcr, in high affinity nitric oxide reduction that protects anaerobic cultures of Escherichia coli against nitrosative stress. Mol Microbiol 100:877–892. doi: 10.1111/mmi.13356. [DOI] [PubMed] [Google Scholar]
- 19.Costa C, Macedo A, Moura I, Moura JJG, Le Gall J, Berlier Y, Liu MY, Payne WJ. 1990. Regulation of the hexaheme nitrite/nitric oxide reductase of Desulfovibrio desulfuricans, Wolinella succinogenes and Escherichia coli: a mass spectrometric study. FEBS Lett 276:67–70. doi: 10.1016/0014-5793(90)80508-G. [DOI] [PubMed] [Google Scholar]
- 20.Poock SR, Leach ER, Moir JWB, Cole JA, Richardson DJ. 2002. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J Biol Chem 277:23664–23669. doi: 10.1074/jbc.M200731200. [DOI] [PubMed] [Google Scholar]
- 21.Gardner AM, Cook MR, Gardner PR. 2010. Nitric-oxide dioxygenase function of human cytoglobin with cellular reductants and in rat hepatocytes. J Biol Chem 285:23850–23857. doi: 10.1074/jbc.M110.132340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott NL, Xu Y, Shen G, Vuletich DA, Falzone CJ, Li Z, Ludwig M, Pond MP, Preimesberger MR, Bryant DA, Lecomte JTJ. 2010. Functional and structural characterization of the 2/2 hemoglobin from Synechococcus sp. PCC 7002. Biochemistry 49:7000–7011. doi: 10.1021/bi100463d. [DOI] [PubMed] [Google Scholar]
- 23.Stern AM, Liu B, Bakken LR, Shapleigh JP, Zhu J. 2013. A novel protein protects bacterial iron-dependent metabolism from nitric oxide. J Bacteriol 195:4702–4708. doi: 10.1128/JB.00836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JLM, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 25.Meyer TE, Tsapin AI, Vandenberghe I, De Smet L, Frishman D, Nealson KH, Cusanovich MA, Van Beeumen JJ. 2004. Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. OMICS 8:57–77. doi: 10.1089/153623104773547499. [DOI] [PubMed] [Google Scholar]
- 26.Gao H, Barua S, Liang Y, Wu L, Dong Y, Reed S, Chen J, Culley D, Kennedy D, Yang Y, He Z, Nealson KH, Fredrickson JK, Tiedje JM, Romine M, Zhou J. 2010. Impacts of Shewanella oneidensis c-type cytochromes on aerobic and anaerobic respiration. Microb Biotechnol 3:455–466. doi: 10.1111/j.1751-7915.2010.00181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin M, Jiang Y, Sun L, Yin J, Fu H, Wu G, Gao H. 2013. Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS One 8:e75610. doi: 10.1371/journal.pone.0075610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford PC. 2010. Reactions of NO and nitrite with heme models and proteins. Inorg Chem 49:6226–6239. doi: 10.1021/ic902073z. [DOI] [PubMed] [Google Scholar]
- 29.Fu H, Chen H, Wang J, Zhou G, Zhang H, Zhang L, Gao H. 2013. Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ Microbiol 15:2198–2212. doi: 10.1111/1462-2920.12091. [DOI] [PubMed] [Google Scholar]
- 30.Jin M, Fu H, Yin J, Yuan J, Gao H. 2016. Molecular underpinnings of nitrite effect on CymA-dependent respiration in Shewanella oneidensis. Front Microbiol 7:1154. doi: 10.3389/fmicb.2016.01154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin M, Zhang Q, Sun Y, Gao H. 2016. NapB in excess inhibits growth of Shewanella oneidensis by dissipating electrons of the quinol pool. Sci Rep 6:37456. doi: 10.1038/srep37456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Fu H, Wang J, Sun L, Jiang Y, Zhang L, Gao H. 2013. Impacts of nitrate and nitrite on physiology of Shewanella oneidensis. PLoS One 8:e62629. doi: 10.1371/journal.pone.0062629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzhugh AL, Keefer LK. 2000. Diazeniumdiolates: pro- and antioxidant applications of the “NONOates.” Free Radic Biol Med 28:1463–1469. doi: 10.1016/S0891-5849(00)00251-3. [DOI] [PubMed] [Google Scholar]
- 34.Price MS, Chao LY, Marletta MA. 2007. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry 46:13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crane BR, Sudhamsu J, Patel BA. 2010. Bacterial nitric oxide synthases. Annu Rev Biochem 79:445–470. doi: 10.1146/annurev-biochem-062608-103436. [DOI] [PubMed] [Google Scholar]
- 36.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol 20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 37.Gusarov I, Starodubtseva M, Wang Z-Q, McQuade L, Lippard SJ, Stuehr DJ, Nudler E. 2008. Bacterial nitric-oxide synthases operate without a dedicated redox partner. J Biol Chem 283:13140–13147. doi: 10.1074/jbc.M710178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao H, Yang ZK, Barua S, Reed SB, Romine MF, Nealson KH, Fredrickson JK, Tiedje JM, Zhou J. 2009. Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J 3:966–976. doi: 10.1038/ismej.2009.40. [DOI] [PubMed] [Google Scholar]
- 39.Gao H, Wang X, Yang ZK, Chen J, Liang Y, Chen H, Palzkill T, Zhou J. 2010. Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS One 5:e15295. doi: 10.1371/journal.pone.0015295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pant K, Bilwes AM, Adak S, Stuehr DJ, Crane BR. 2002. Structure of a nitric oxide synthase heme protein from Bacillus subtilis. Biochemistry 41:11071–11079. doi: 10.1021/bi0263715. [DOI] [PubMed] [Google Scholar]
- 41.Fu H, Jin M, Ju L, Mao Y, Gao H. 2014. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ Microbiol 16:3181–3195. doi: 10.1111/1462-2920.12457. [DOI] [PubMed] [Google Scholar]
- 42.Brettar I, Christen R, Höfle MG. 2002. Shewanella denitrificans sp. nov., a vigorously denitrifying bacterium isolated from the oxic-anoxic interface of the Gotland Deep in the central Baltic Sea. Inter J Syst Evol Microbiol 52:2211–2217. doi: 10.1099/00207713-52-6-2211. [DOI] [PubMed] [Google Scholar]
- 43.Plate L, Marletta MA. 2012. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol Cell 46:449–460. doi: 10.1016/j.molcel.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong Y, Wang J, Fu H, Zhou G, Shi M, Gao H. 2012. A Crp-dependent two-component system regulates nitrate and nitrite respiration in Shewanella oneidensis. PLoS One 7:e51643. doi: 10.1371/journal.pone.0051643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vicente JB, Carrondo MA, Teixeira M, Frazão C. 2007. Flavodiiron proteins: nitric oxide and/or oxygen reductases. In Messerschmidt A. (ed), Handbook of metalloproteins. John Wiley & Sons, Inc.,, Hoboken, NJ. doi: 10.1002/0470028637.met218. [DOI] [Google Scholar]
- 46.Vuletich D, Lecomte JJ. 2006. A phylogenetic and structural analysis of truncated hemoglobins. J Mol Evol 62:196–210. doi: 10.1007/s00239-005-0077-4. [DOI] [PubMed] [Google Scholar]
- 47.Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol 1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo Q, Dong Y, Chen H, Gao H. 2013. Mislocalization of Rieske protein PetA predominantly accounts for the aerobic growth defect of tat mutants in Shewanella oneidensis. PLoS One 8:e62064. doi: 10.1371/journal.pone.0062064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu H, Jin M, Wan F, Gao H. 2015. Shewanella oneidensis cytochrome c maturation component CcmI is essential for heme attachment at the non-canonical motif of nitrite reductase NrfA. Mol Microbiol 95:410–425. doi: 10.1111/mmi.12865. [DOI] [PubMed] [Google Scholar]
- 50.Yin J, Jin M, Zhang H, Ju L, Zhang L, Gao H. 2015. Regulation of nitrite resistance of the cytochrome cbb3 oxidase by cytochrome c ScyA in Shewanella oneidensis. Microbiologyopen 4:84–99. doi: 10.1002/mbo3.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou G, Yin J, Chen H, Hua Y, Sun L, Gao H. 2013. Combined effect of loss of the caa3 oxidase and Crp regulation drives Shewanella to thrive in redox-stratified environments. ISME J 7:1752–1763. doi: 10.1038/ismej.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marrs B, Gest H. 1973. Genetic mutations affecting the respiratory electron-transport system of the photosynthetic bacterium Rhodopseudomonas capsulata. J Bacteriol 114:1045–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan F, Shi M, Gao H. 2017. Loss of OxyR reduces efficacy of oxygen respiration in Shewanella oneidensis. Sci Rep 7:42609. doi: 10.1038/srep42609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardner PR, Costantino G, Szabó C, Salzman AL. 1997. Nitric oxide sensitivity of the aconitases. J Biol Chem 272:25071–25076. doi: 10.1074/jbc.272.40.25071. [DOI] [PubMed] [Google Scholar]
- 55.Pereira MM, Sousa FL, Veríssimo AF, Teixeira M. 2008. Looking for the minimum common denominator in haem-copper oxygen reductases: towards a unified catalytic mechanism. Biochim Biophys Acta 1777:929–934. doi: 10.1016/j.bbabio.2008.05.441. [DOI] [PubMed] [Google Scholar]
- 56.Sarti P, Forte E, Mastronicola D, Giuffrè A, Arese M. 2012. Cytochrome c oxidase and nitric oxide in action: molecular mechanisms and pathophysiological implications. Biochim Biophys Acta 1817:610–619. doi: 10.1016/j.bbabio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Einsle O. 2011. Structure and function of formate-dependent cytochrome c nitrite reductase, NrfA. Methods Enzymol 496:399–422. doi: 10.1016/B978-0-12-386489-5.00016-6. [DOI] [PubMed] [Google Scholar]
- 58.Clarke TA, Kemp GL, Van Wonderen JH, Doyle RM, Cole JA, Tovell N, Cheesman MR, Butt JN, Richardson DJ, Hemmings AM. 2008. Role of a conserved glutamine residue in tuning the catalytic activity of Escherichia coli cytochrome c nitrite reductase. Biochemistry 47:3789–3799. doi: 10.1021/bi702175w. [DOI] [PubMed] [Google Scholar]
- 59.Kern M, Volz J, Simon J. 2011. The oxidative and nitrosative stress defence network of Wolinella succinogenes: cytochrome c nitrite reductase mediates the stress response to nitrite, nitric oxide, hydroxylamine and hydrogen peroxide. Environ Microbiol 13:2478–2494. doi: 10.1111/j.1462-2920.2011.02520.x. [DOI] [PubMed] [Google Scholar]
- 60.Figueiredo MCO, Lobo SAL, Sousa SH, Pereira FP, Wall JD, Nobre LS, Saraiva LM. 2013. Hybrid cluster proteins and flavodiiron proteins afford protection to Desulfovibrio vulgaris upon macrophage infection. J Bacteriol 195:2684–2690. doi: 10.1128/JB.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi M, Wan F, Mao Y, Gao H. 2015. Unraveling the mechanism for the viability deficiency of Shewanella oneidensis oxyR null mutant. J Bacteriol 197:2179–2189. doi: 10.1128/JB.00154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miranda KM, Espey MG, Wink DA. 2001. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5:62–71. doi: 10.1006/niox.2000.0319. [DOI] [PubMed] [Google Scholar]
- 63.Wu L, Wang J, Tang P, Chen H, Gao H. 2011. Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS One 6:e21479. doi: 10.1371/journal.pone.0021479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi M, Gao T, Ju L, Yao Y, Gao H. 2014. Effects of FlrBC on flagellar biosynthesis of Shewanella oneidensis. Mol Microbiol 93:1269–1283. doi: 10.1111/mmi.12731. [DOI] [PubMed] [Google Scholar]
- 65.Yuan J, Wei B, Shi M, Gao H. 2011. Functional assessment of EnvZ/OmpR two-component system in Shewanella oneidensis. PLoS One 6:e23701. doi: 10.1371/journal.pone.0023701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen H, Luo Q, Yin J, Gao T, Gao H. 2015. Evidence for the requirement of CydX in function but not assembly of the cytochrome bd oxidase in Shewanella oneidensis. Biochim Biophys Acta 1850:318–328. doi: 10.1016/j.bbagen.2014.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.