FIGURE 3.
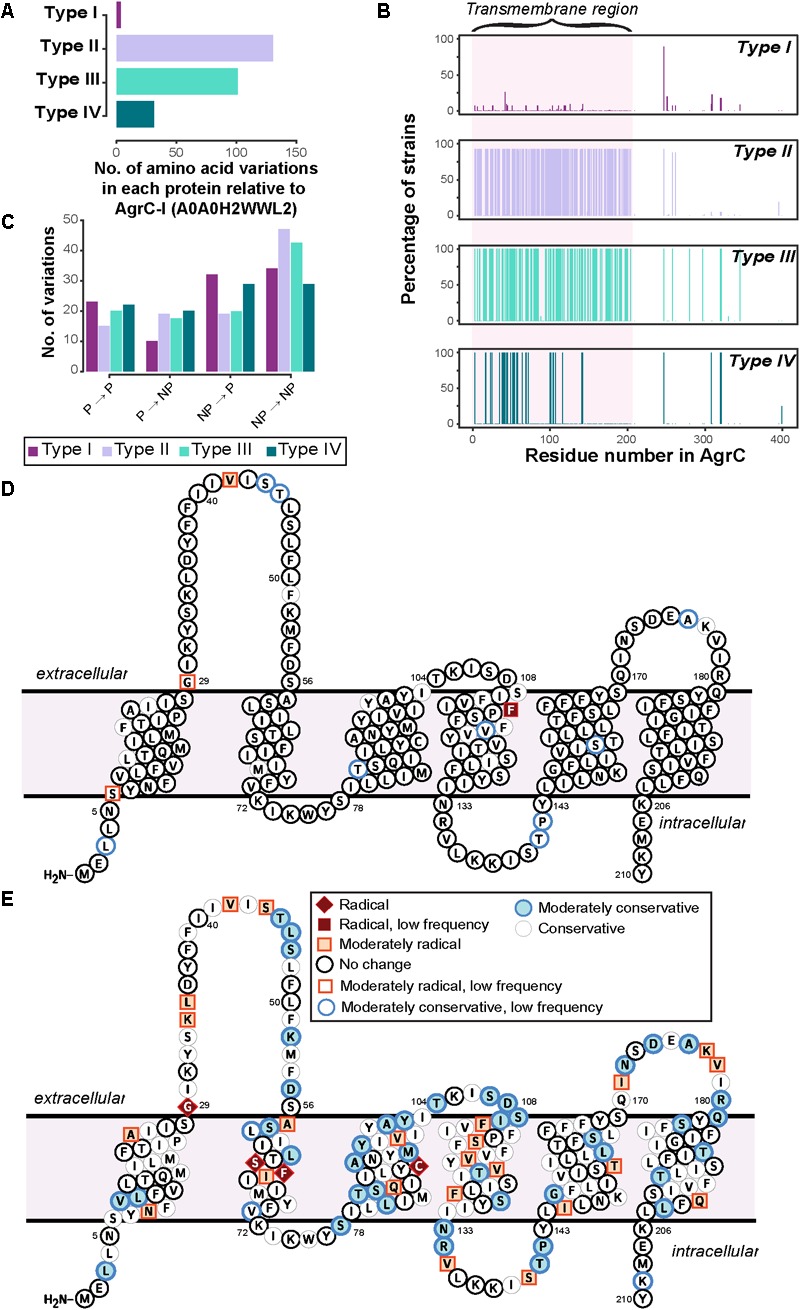
Sequence divergence in AgrC of four agr types. (A) The number of amino acid variations in each protein of agr types. Type I shows the least variation, as the reference strain used for the analysis is of type I. AgrC sequence of type II and type III have the highest number of amino acid variations. (B) The percent of strains having a variation in that residue number. Around 92 and 100% of strains of type II and type III, respectively, have variations mostly in the first 200 residues of AgrC protein. (C) Properties of amino acid variations in each type show that the majority of variations are between non-polar amino acids in type II and type III, meaning there is a much higher divergence in the transmembrane region. Abbreviations: NP, non-polar; P, Polar. (D,E): specific amino acid divergence between type I and type II. The predicted transmembrane topology of AgrC-I reference sequence from CCTOP with highlighted amino acid residues that tend to diverge in (D) type I strains and (E) type II strains, from this study. The color coding is per biochemical and biophysical property of the amino acid residue mutated. Radical, moderate, and conservative is the nomenclature given in the order of amino acid divergence. Radical is a vast difference between amino acid properties and could change the properties of the protein. Conservative is an amino acid substitution which can be tolerated by the protein. Moderate is a biochemically moderate change in the amino acid substitution. Low frequency: amino acid variations that occurred only in very few strains.
