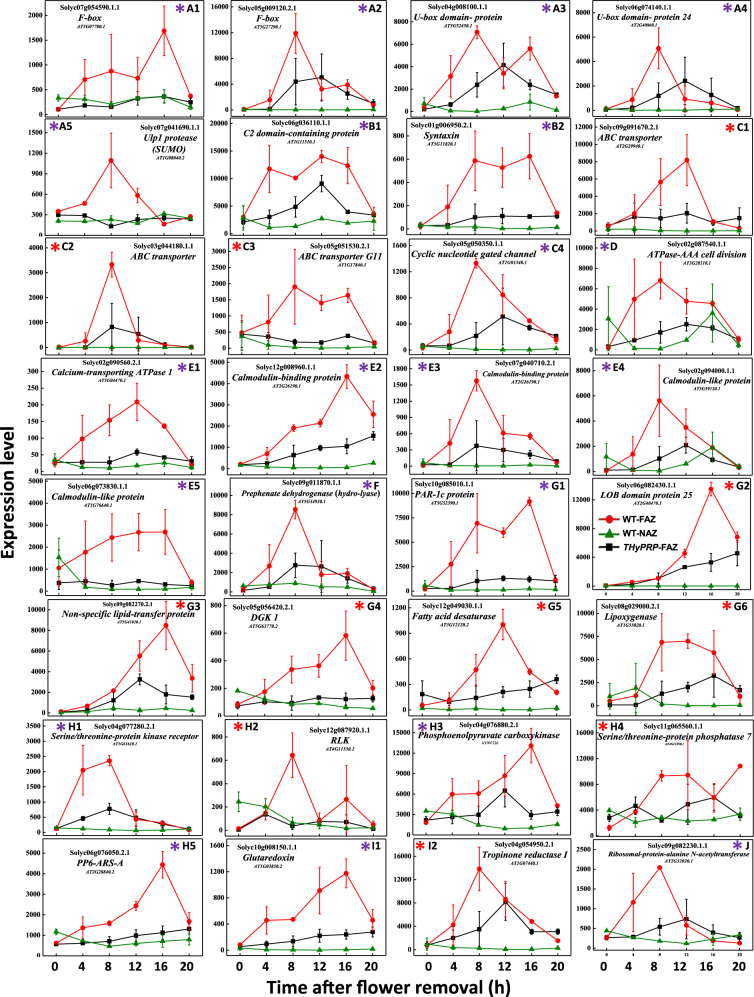
Fig. 6. Effect of antisense silencing of THyPRP on the kinetics of changes in array-measured expression levels of genes that were specifically upregulated in the WT FAZ at various time points after flower removal: early—4 h (blue*) or 8 h (red*) or late 12–20 h (green*).
TAPG4::antisense THyPRP silenced line 11/ generation T4 was used. Expression levels were measured for protein degradation–ubiquitin-related genes—F-box or U-box (a1–a5); genes related to transporters of macromolecules—C2 domain-containing protein (b1) and Syntaxin (b2); ABC transporter genes (c1–c3); Cyclic nucleotide-gated ion channels (c4); ATPase-AAA cell division (d); Calcium-transporting ATPase1 (e1); Calmodulin-binding protein genes (e2, e3); Calmodulin-like protein genes (e4–e5); Prephenate dehydrogenase hydrolyase (f); Lateral organ boundaries (LOB) lipids and wax-related genes—photoassimilate-responsive-1c (PAR-1c protein) (g1); LOB-domain protein25 (g2); Non-specific lipid-transfer protein (g3); Diacylglycerol kinase1 (DGK1) (g4); Fatty acid desaturase (g5); Lipoxygenase (g6); receptor kinase and protein phosphatase-related genes—Serine/threonine-protein kinase receptor (h1); Receptor-Like protein Kinase (RLK) (h2); Phosphoenolpyruvate carboxykinase (h3); Serine/threonine-protein phosphatase7 (h4); Serine/threonine-Protein Phosphatase6 regulatory Ankyrin Repeat Subunit A (PP6-ARS-A) (h5); Redox regulation genes—Glutaredoxin (i1); Tropinone reductase1 (i2); and Ribosomal-protein-alanine N-acetyl-transferase (j). Transcript identities are indicated in the graphs by their gene ID and their Arabidopsis (At) gene number, and/or their nucleotide accession number. The results are means of two biological independent replicates ± SD