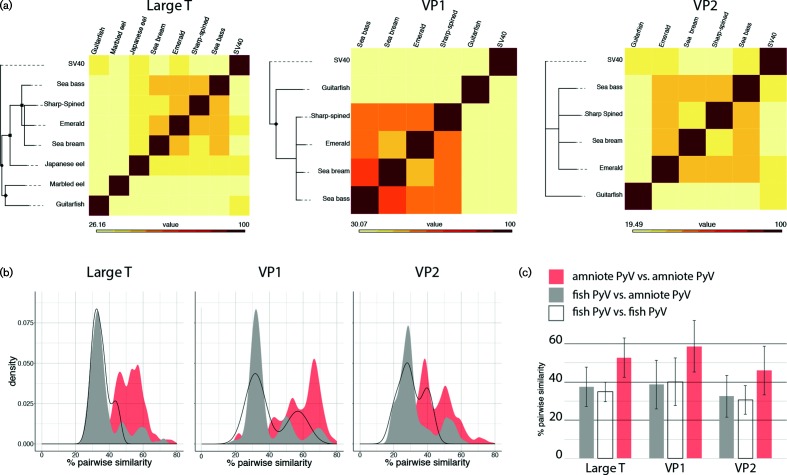
Fig. 2.
Fish viral proteins are evolutionarily divergent (a) maximum likelihood phylogenetic trees of individual protein trees. English host names are used to refer to the fish-associated PyVs. Trees were rooted using SV40, and the remaining (n=114) amniote PyVs were removed to improve visualization (complete trees available in Fig. S2). Symbols indicate bootstrap support (square=100; diamond >90; circle >80). Nodes with bootstrap support less than 70 % were collapsed. Heat maps to the right of the tree show the pairwise sequence similarity. (b) Graphs comparing the percentage pairwise similarity of different PyV proteins. The black line shows the distribution of pairwise comparisons of fish PyVs. The grey plot represents comparisons between fish and amniote PyV, while the red plots show comparisons between amniote PyV. This data shows that amniote PyVs are more similar to other amniote PyVs than fish PyVs are to each other. (c) Bar graphs showing average and sd of the curves in (b).