Fig. 1.
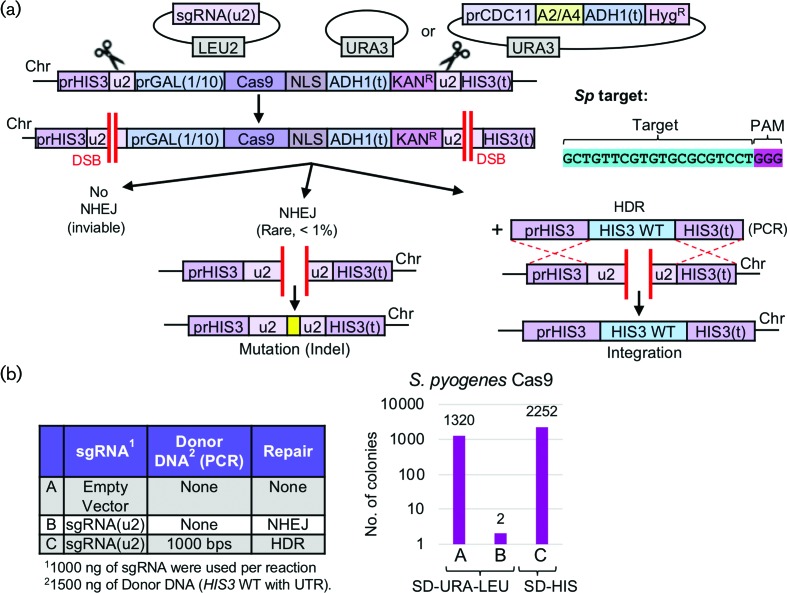
CRISPR/Cas9 editing of a haploid yeast genome using S. pyogenes Cas9 and an artificial target system. (a) Schematic of the yeast Cas9 expression platform at the endogenous HIS3 locus. The Cas9 gene is under control of the inducible GAL1/10 promoter and the locus is marked with the KanR cassette. The entire expression module is flanked by two identical artificial [u2] sites (32 inserted base pairs including a PAM sequence), as previously described. A high-copy LEU2-marked plasmid harbours the sgRNA[u2] cassette whereas a URA3-based plasmid is also present (empty or expressing an anti-CRISPR gene). S. pyogenes Cas9 was synthesized with a yeast codon bias and integrated. (b) Yeast strain GFY-2383 harbouring an empty URA3-based plasmid (pRS316) was cultured in pre-induction medium (raffinose/sucrose mixture) overnight at 30 °C, back-diluted to an OD600 of approximately 0.35 in YPGal medium and grown for 4.5 additional hours. Cells were harvested, transformed with the equimolar amounts of an empty vector control (A, pRS425) or sgRNA[u2] plasmid (B, pGF-V809), recovered overnight in fresh YPGal medium and plated onto SD-URA-LEU selection plates. In one sample, the guide RNA plasmid was co-transformed with a PCR fragment (C, WT HIS3 ORF with 1000 bp of flanking 5′ and 3′ UTR). The total number of surviving colonies was quantified and graphed on a log10 scale.