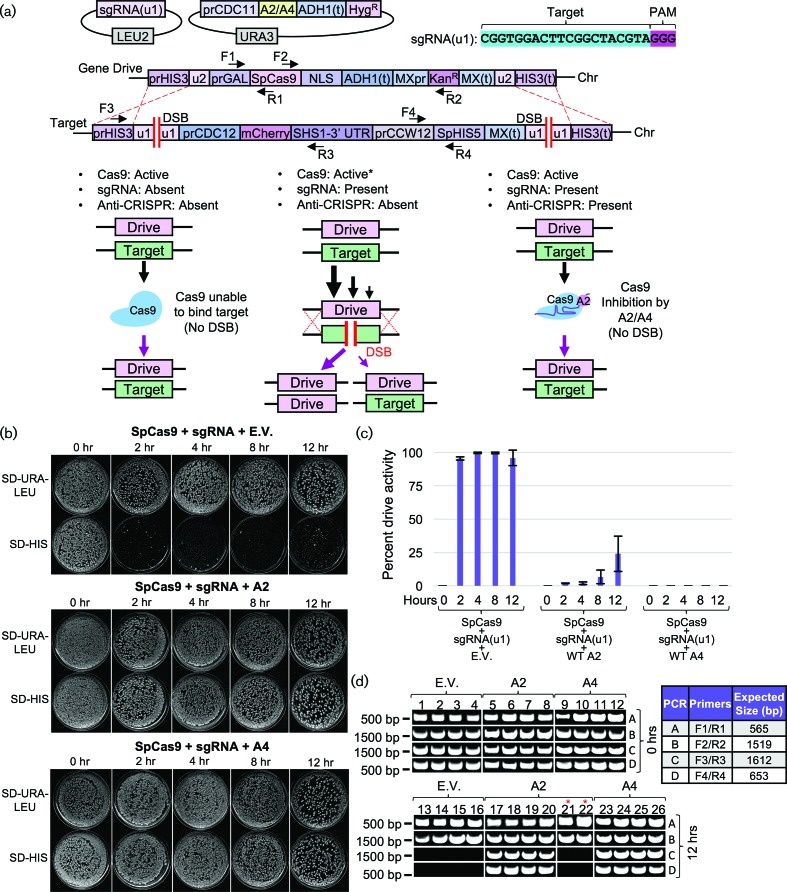
Fig. 4.
AcrIIA2 and AcrIIA4 can inhibit CRISPR-based gene drives in yeast. (a) Design of an artificial gene drive system in S. cerevisiae. The haploid ‘gene drive’ strain (harbouring inducible S. pyogenes Cas9) was built in MATa yeast (GFY-2383) and included (i) a LEU2-based high copy plasmid with the sgRNA[u1] cassette and (ii) the URA3-based CEN plasmid expressing untagged AcrIIA2 or AcrIIA4 (top). Two nearly isogenic ‘target’ strains were built in MATα yeast (GFY-3206 and GFY-3207) containing an artificial target gene and selectable HIS5 marker (from S. pombe). The entire construct was flanked with two [u1] artificial target sites. Middle, three scenarios are depicted involving (i) a fully active gene drive, (ii) partially active drive activity and (iii) fully inhibited drive activity due to the presence of the anti-CRISPR proteins. (b) Activation of an artificial gene drive system in yeast. First, yeast were transformed with (i) either an empty vector (pRS316) or plasmid expressing AcrIIA2 (pGF-IVL1336) or AcrIIA4 (pGF-IVL1337) and (ii) the sgRNA[u2] plasmid (pGF-V1220), and mated to the target strains (pGF-3206 and pGF-3207) on rich medium (containing dextrose) for 24 h at 30 °C. Second, diploid yeast were obtained by velvet transfer of all colonies to SD-URA-LEU-HIS medium for two consecutive rounds of selection. Third, cultures of pre-induction medium (raffinose/sucrose lacking leucine and lacking uracil) were grown overnight, back-diluted into YPGal and cultured for between 0 and 12 h. Fourth, cells were harvested, washed and diluted to approximately 500–1000 cells per plate (SD-URA-LEU medium) and grown for 2–3 days. Fifth, yeasts were transferred by velvet to an identical plate type and SD-HIS medium for an additional 24 h incubation before imaging. Representative plates for each time point are illustrated. (c) The total number of surviving colonies was quantified for each plate type in duplicate. ‘Gene drive activity’ was illustrated as the proportion of sampled colonies (n=100–300 colonies per plate) sensitive to the SD-HIS condition (e.g. 99 % of colonies present on SD-URA-LEU plate but absent on the SD-HIS plate corresponds to 99 % gene drive activity). (d) Molecular analysis of diploid yeast following gene drive activation. Clonal isolates were obtained from the 0 and 12 h time points (SD-URA-LEU plate), chromosomal DNA was purified and PCRs were performed on the diploid genomes. Primer combinations and the expected fragment sizes (right) are illustrated in the gene drive schematic (a). Four representative isolates from each genotype are illustrated – for the gene drive containing AcrIIA2, two isolates displaying no growth on SD-HIS were also tested (red asterisk).