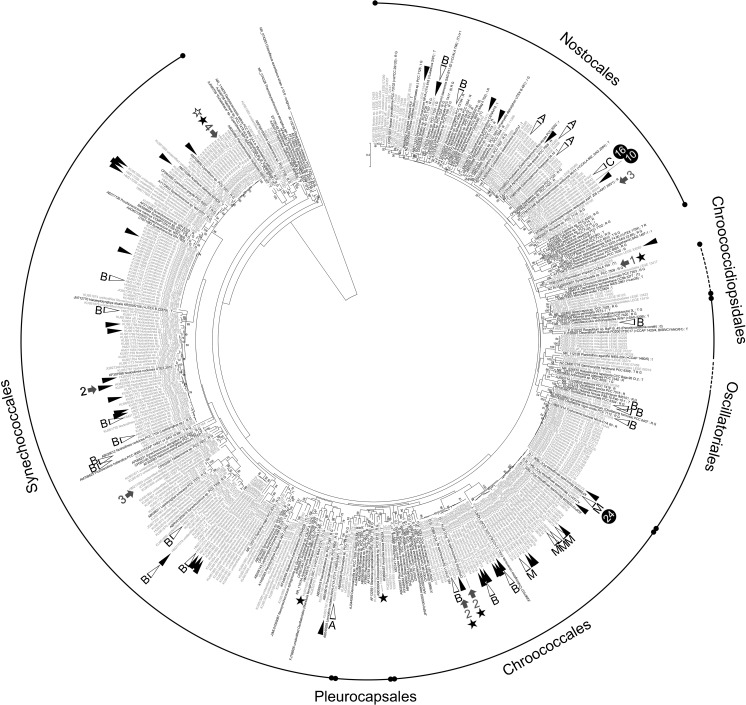
Fig. 3.
Circular ML tree (− lnl = 25,944.6863) of 16S rRNA gene sequences illustrating the phylogenetic diversity of LEGE CC strains (in gray), their placement at the order level, and some traits or information relevant for biotechnological purposes. One hundred and fifty-two sequences from reference material (Ramos et al. 2017) were included to disclose the cyanobacterial “Tree of Life” (T or t stand for type strains designated as representing type species, R or r for reference strains sensu Bergey’s Manual (Castenholz et al. 2001); and G for genome sequences available; see also Material and methods section for details). Accession numbers for all sequences are shown. Only bootstrap support values over 50% are given. Black arrowheads indicate strains capable of producing good amounts of EPSs. White arrowheads denote strains producing the following cyanotoxins, as demonstrated by analytical chemistry methods: A anatoxin-a, e.g., Osswald et al. (2009); B BMAA (Cianca et al. 2012); C cylindrospermopsin, e.g., Saker and Eaglesham (1999); and M microcystin, e.g., Vasconcelos et al. (1995). Arrows point to strains used to isolate and elucidate the structure of the following secondary metabolites: 1 hierridin B (Leão et al. 2013b), 2 portoamides (Leão et al. 2010), 3 bartolosides (Leão et al. 2015; Afonso et al. 2016), and 4 dehydroabietic acid (Costa et al. 2016). Black stars indicate strains having (or soon will have) their genome sequenced, and the white star stands for a strain that has a submitted patent application. Black circles and numbers within refer to highly used strains and to the number of times they appear in the literature, respectively