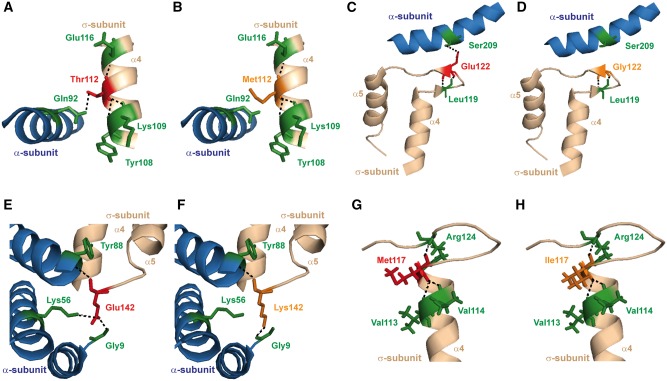
Figure 3.
Structural characterization of the AP2σ variants within the AP2σ α4-α5 helices encoded by AP2S1 exon 5. (A) Structural model of the α4-helix of the AP2σ subunit (shown in light brown) with the adjacent α5-helix of the AP2α subunit (shown in blue). The Thr112 residue forms a polar contact with the Tyr108, Lys109 and Glu116 residues on the α4-helix of AP2σ, and the Gln92 residue on the α5-helix of the AP2α subunit. (B) Mutation of the Thr112 residue to Met112 leads to loss of the polar contact with Gln92, and therefore may impair AP2σ–AP2α subunit interactions. (C) Structural model of the α4–α5 helices of the AP2σ subunit (shown in light brown) with the adjacent α12-helix of the AP2α subunit (shown in blue). The Glu122 residue is located within the α4–α5 loop, and forms polar contacts with Leu119 of the AP2σ subunit, and Ser209 in the α12-helix of the AP2α subunit. (D) Mutation of residue Glu122 to Gly122 results in loss of the contact with Ser209, and thus the Gly122 variant may impair AP2σ–AP2α subunit interactions. (E) Structural model of the α4 and α5 helices of AP2σ (shown in light brown) and the AP2α subunit (shown in blue). The Glu142 residue is located at the end of the α4-helix of the AP2σ subunit and forms polar contacts with Gly9, Lys56 and Tyr88 of the AP2α subunit. (F) Mutation of the Glu142 residue to Lys142 disrupts the polar contact with Lys56, and thus the Lys142 variant may impair AP2σ–AP2α subunit interactions. (G) Structural model of the α4 helix of AP2σ (shown in light brown) with the Met117 residue indicated in red. The Met117 residue forms polar contacts with Val113, Val114 and Arg124. (H) Mutation of the Met117 residue to Ile117 is not predicted to alter residue hydrophobicity and disrupt these polar contacts.