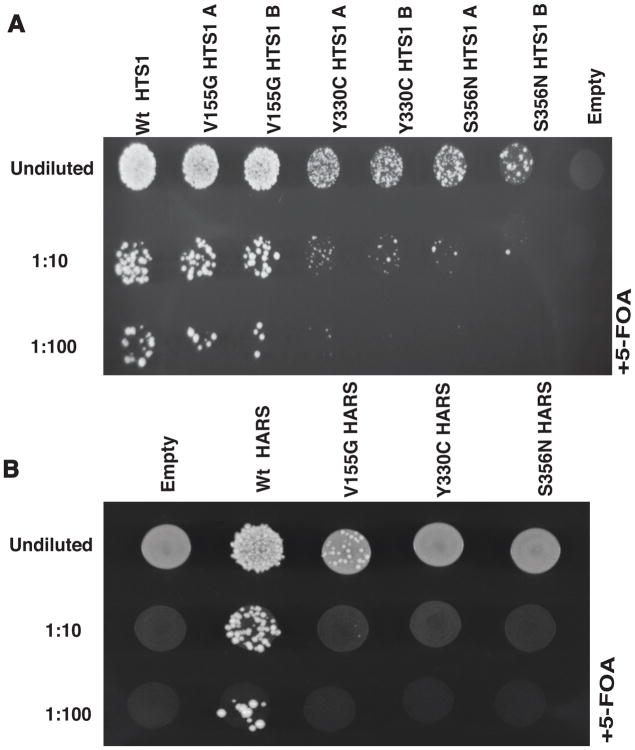
Figure 4. Neuropathy-associated HARS mutations result in loss-of-function in vivo.
(A) Yeast complementation analysis of HARS variants. Haploid ΔHTS1 yeast strains were transformed with a vector containing no insert (‘Empty’) or an insert to express wild-type, p.Val155Gly, p.Tyr330Cys, or p.Ser356Asn HTS1. Two colonies (indicated by ‘A’ and ‘B’) from transformations with p.Val155Gly, p.Tyr330Cys, or p.Ser356Asn HTS1 are shown. Resulting colonies (undiluted, diluted 1:10, or diluted 1:100) were grown on agar plates containing complete media with 0.1% 5-FOA. Note the severe depletion of growth associated with p.Tyr330Cys and p.Ser356Asn HTS1 in the 1:10 and 1:100 dilutions. (B) Similar yeast complementation assays as describe in A using the human HARS open-reading frame. Haploid ΔHTS1 yeast strains were transformed with a vector containing no insert (‘Empty’) or an insert to express wild-type, p.Val155Gly, p.Tyr330Cys, or p.Ser356Asn HARS. After transformations, colonies (undiluted, diluted 1:10, or diluted 1:100) were grown on agar plates containing complete media with 0.1% 5-FOA. Note the severe depletion of growth associated with p.Val155Gly HARS in the undiluted sample and the lack of growth in the 1:10 and 1:100 dilutions.