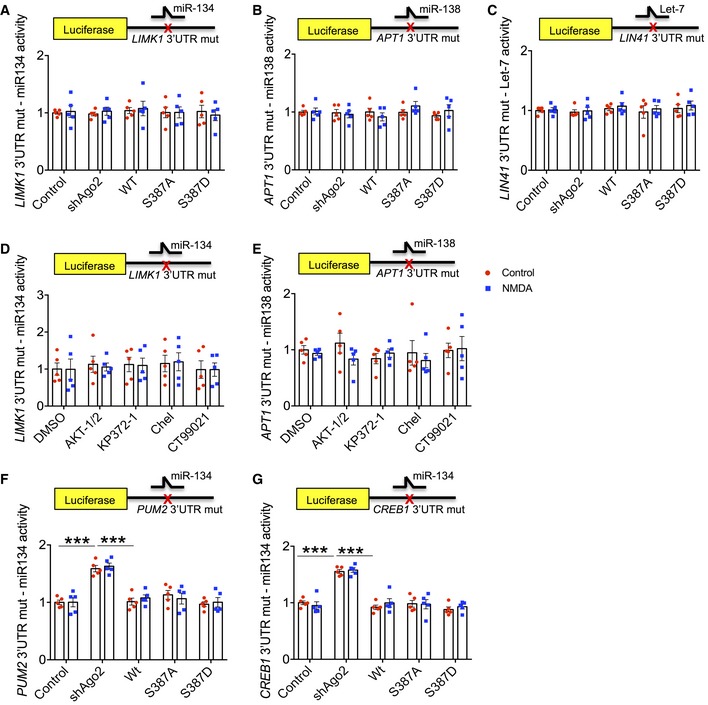
The effects on luciferase reporters incorporating
LIMK1,
APT1 and
LIN41 3′UTRs seen in Fig
5 are fully explained by regulation via miR‐134, miR‐138 and Let‐7, respectively. Cultured cortical neurons transfected with Ago2 molecular replacement constructs as well as
Renilla luciferase and
Firefly luciferase reporters containing LIMK1 (A, D), APT1 (B, E) or LIN41 (C) 3′UTRs containing mutations in the seed regions for miR‐134, miR‐138 and Let‐7, respectively, were treated with NMDA or vehicle for 3 min. Ten minutes after NMDA washout, lysates were prepared for dual‐luciferase assays. For (D, E), cultures were also treated with kinase inhibitors as shown 20 min before NMDA or vehicle. Error bars are SEM.