Figure 1. Hfq is required for correct processing and folding of 16S rRNA .
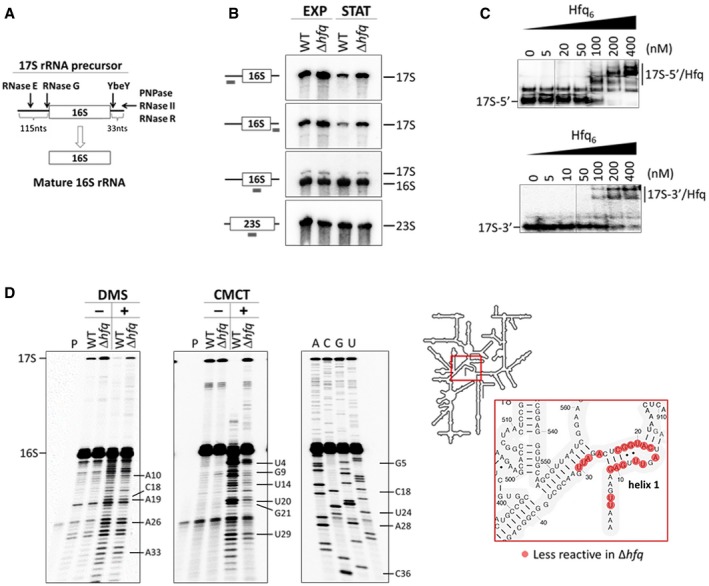
- Schematic representation of the RNase‐mediated processing of the 17S rRNA precursor into mature 16S rRNA.
- Northern blot analysis of total RNA extracted from cells in exponential (EXP) or stationary (STAT) growth phase. Samples were fractionated on a 4% polyacrylamide/7 M urea gel. A scheme of the probes binding to the rRNA sequence is displayed on the side.
- Electrophoretic mobility shift assays of Hfq binding to the 5′ and 3′ extremities of the 17S rRNA. Increasing amounts of Hfq hexamer were mixed with a constant amount of the specific 17S‐flanking sequences and resolved on a 6% (top panel) or 8% (bottom panel) native polyacrylamide gel.
- DMS and CMCT accessibility probing of the 16S rRNA. Reverse‐transcribed cDNA was fractionated on an 10% polyacrylamide/7 M urea gel. Residues with altered reactivities in the Δhfq mutant are indicated. The inset depicts the analyzed region of the 16S rRNA.
Source data are available online for this figure.
