Figure EV3. Cumulative metagene profile and coverage profiles of selected downregulated genes in the wild‐type and Δhfq strain obtained by ribosome profiling.
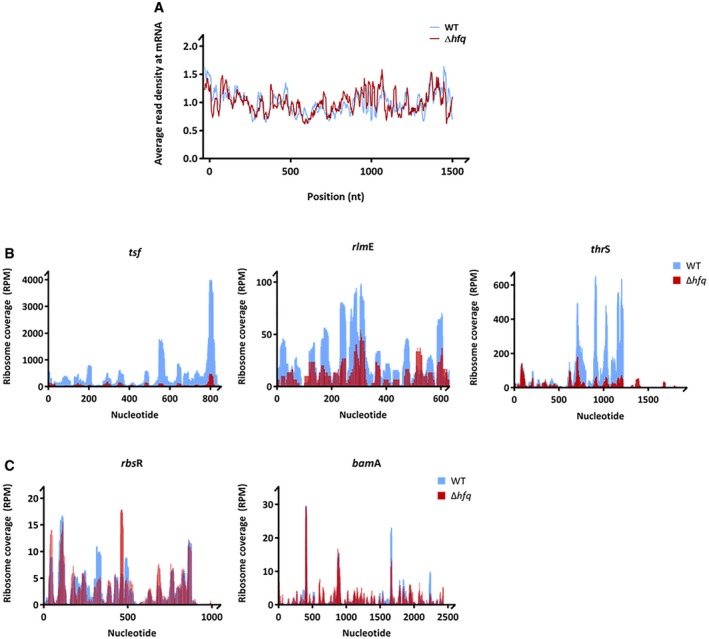
- Cumulative metagene profile of the read density as a function of position for RPFs. The expressed genes were individually normalized, aligned at the start codon, and averaged with equal weight. 1,075 and 1,231 genes from wild‐type and Δhfq strains, respectively, were considered in the analysis.
- Coverage profiles of selected downregulated genes representative of gene categories affected by Hfq deletion (Fig 4D). Elongation factor‐Ts (tsf), 23S rRNA 2′‐O‐ribose U2552 methyltransferase (rlmE) are included in the GO term “translation and ribosome” and threonine‐tRNA ligase (thrS) in the GO term “amino acid biosynthesis”.
- Coverage profiles of exemplified genes (bamA and rbsR) whose expression remained unchanged upon hfq deletion. BamA is an outer membrane protein assembly factor, and RbsR is a transcriptional factor of the operon involved in ribose catabolism and transport.
