Figure 5. SUP interacts with PcG proteins CLF and TFL2.
-
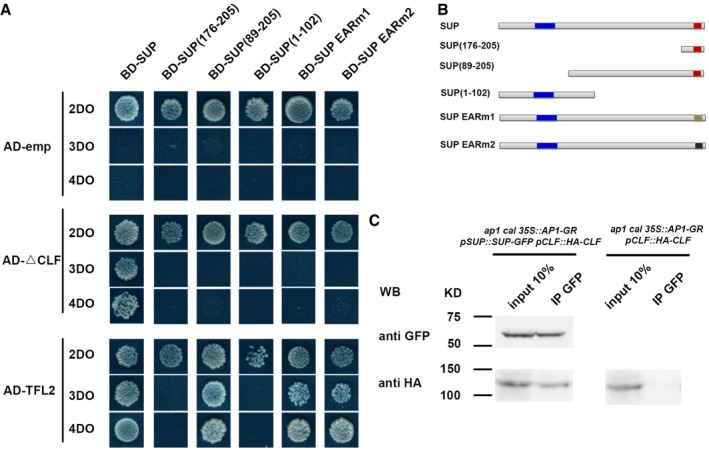
A, BThe yeast two‐hybrid assay using a series of truncated SUP proteins with CLF and TFL2. Full‐length, three truncated SUP proteins and SUP proteins with the two versions of the abolished EAR motif (EARm1 and EARm2) were fused to a GAL4 DNA‐binding domain (BD). Schematic structures of the full‐length, truncated, or mutated SUP protein are shown in (B). The blue region indicates the zinc finger domain; the red regions indicate an intact EAR motif; and beige and black regions indicate mutated EAR motif, respectively. The truncated CLF without its C‐terminal SET domain (CLF) and the full‐length TFL2 was fused to the GAL4 activation domain (AD). Yeast colonies harboring these fusion constructs and/or empty vectors as indicated were grown on selective media of 2DO, 3DO, and 4DO. For CLF, yeast growth was only detected when the combination of the full‐length SUP and the truncated CLF was co‐transformed. None of the truncated SUP proteins or SUP proteins with the mutated EAF motif interacted with the truncated CLF. For TFL2, a short domain of SUP (amino acids 89–205) was sufficient for the interaction, and the mutation of EAR motifs did not affect the interaction with TFL2.
-
CInteraction between SUP and CLF as determined by co‐IP. Using stage 4 floral buds of ap1 cal p35S::AP1‐GR pSUP::SUP‐GFP pCLF::HA‐CLF or of ap1 cal p35S:: AP1‐GR pCLF::HA‐CLF plants, total proteins (input) were subjected to immunoprecipitation with anti‐GFP‐conjugated beads. Immunoblotting analysis with another anti‐GFP and anti‐HA antibody were performed to detect SUP‐GFP (to test pull‐down efficiency) and HA‐CLF (protein interactions). Only in the samples containing both SUP‐GFP and HA‐CLF, HA‐CLF was pull‐down together with anti‐GFP antibody. Molecular mass of the protein ladder is indicated in kilodaltons (KD).
Source data are available online for this figure.
