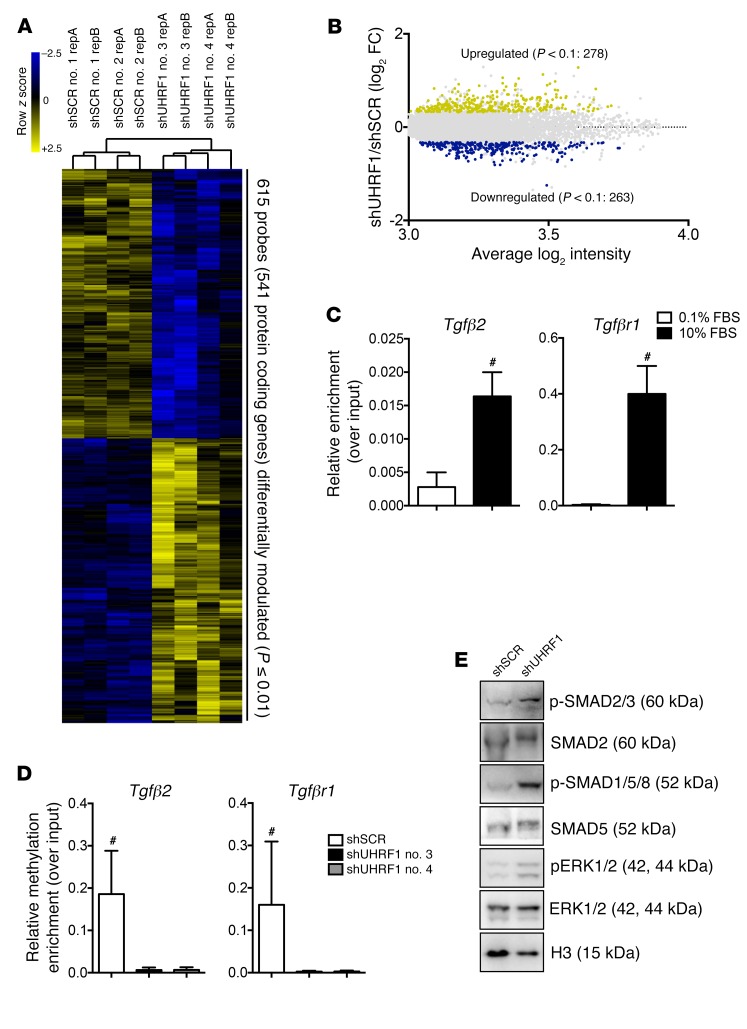
Figure 8. Effects of Uhrf1 absence in vitro.
(A) Hierarchical clustering heat map of 615 probes differentially expressed between shUHRF1 and shSCR (P ≤ 0.1, –1.3 ≤ fold-change ≥ 1.3). (B) Dot plot, with blue dots representing downregulated protein-coding genes (263) and yellow dots representing upregulated protein-coding genes (278) in shUHRF1 and shSCR cells. (C) ChIP assay showing UHRF1 enrichment at the Tgfβ2 and Tgfβr1 promoters in proliferating VSMCs (cells cultured with 10% FBS). Data are presented as mean relative enrichment over input ± SD of 3 biological repeats. (D) Methyl ChIP assay showing reduced methylation at the Tgfβ2 and Tgfβr1 promoters in the absence of UHRF1 (cells cultured with 10% FBS). Data are presented as mean relative enrichment over input ± SD of 3 biological repeats. (E) Representative Western blots showing activation of the canonical TGF-β pathway in Uhrf1-silenced VSMCs. Error bars indicate SD. To compare means, we used unpaired 2-tailed Student’s t test in C and 1-way ANOVA with Tukey’s multiple comparisons t test in D. #P < 0.05 (P value is only adjusted in D).