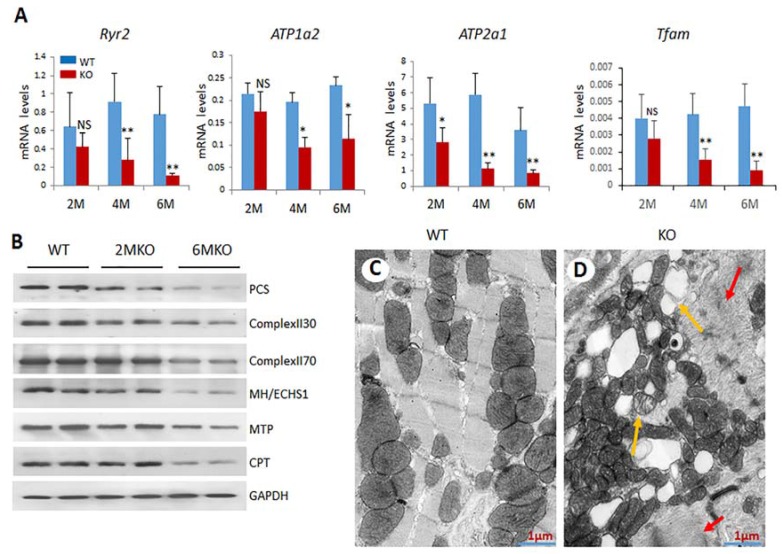
Figure 3.
csPIMT−/− hearts show significant mitochondrial damage. (A) Quantification of mRNA levels of Atp1a2, Atp2a1, Ryr2 and Tfam genes. Each group was analyzed using 5 different mice (assayed individually) and the values are expressed as the mean ± SD. * p < 0.05, ** p < 001, NS: not significant; (B) Western blot showing protein levels for PCS (palmitoyl-CoA synthetase; 62 Kda), complex II30 and 70, MH/ECHS1 (mitochondrial enoyl-CoA hydratase; 31 Kda), MTP (mitochondrial trifunctional protein; 100 Kda) and CPT1α (carnitine palmitoyltransferase; 88 Kda)). The protein extracts were prepared from 5 hearts pooled together. The protein expression of each gene was normalized to GAPDH. Percent decrease as compared WT controls were as follows: PCS, 67%; complex II30 and 70, 28% and 38%; MTP, 26%, and CPT, 42%; (C,D) display the electron micrographs of 6-month csPIMTfl/fl and csPIMT−/− mouse hearts. Red arrows in D indicate abnormal sarcomeres and H zone absent. Yellow arrows point to lipid droplets and damage in mitochondria.