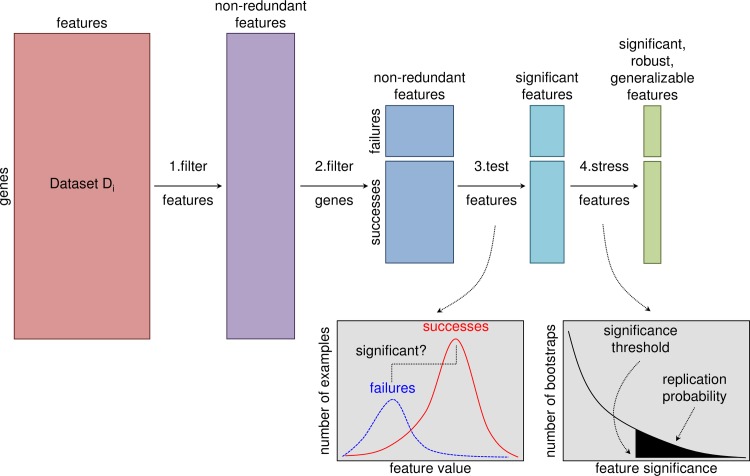
Fig 1. Feature selection pipeline.
Each dataset took the form of a matrix with genes labeling the rows and features labeling the columns. We appended the mean and standard deviation computed across all features as two additional features. Step 1: We filtered the columns to eliminate redundant features, replacing each group of correlated features with the group average feature, where a group was defined as features with squared pair-wise correlation coefficient r2 ≥ 0.5. If the dataset mean feature was included in a group of correlated features, we replaced the group with the dataset mean. Step 2: We filtered the rows for targets with clinical trial outcomes of interest: targets of selective drugs approved for non-cancer indications (successes) and targets of selective drug candidates that failed in phase III clinical trials for non-cancer indications (failures). Step 3: We tested the significance of each feature as an indicator of success or failure using permutation tests to quantify the significance of the difference between the means of the successful and failed targets. We corrected for multiple hypothesis testing using the Benjamini-Yekutieli method to control the false discovery rate at 0.05 within each dataset. Step 4: We “stressed” the significant features with additional tests to assess their robustness and generalizability. For example, we used bootstrapping to estimate probabilities that the significance findings will replicate on similar sets of targets.