Fig. 2. Structure-based alignments of various LsrK and HPr sequences.
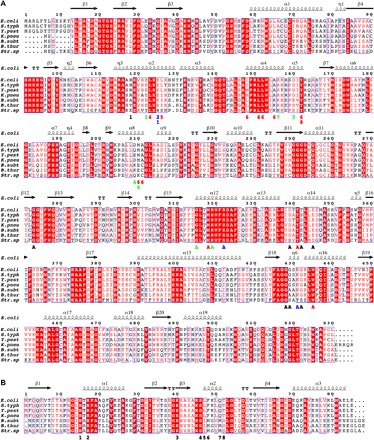
Various LsrK (A) and HPr (B) sequences of Gram-negative [Escherichia coli (E. coli), Salmonella typhimurium LT2 (S. typh), Yersinia pestis (Y. pest), and Klebsiella pneumoniae subsp. pneumoniae (K. pneu)] and Gram-positive [Bacillus subtilis (B. subt), Bacillus thuringiensis (B. thur), and Streptococcus sp. oral taxon 056 (Str. sp)] bacteria were aligned on the basis of the determined structure of the HisLsrK/HPr/ATP complex. The E. coli LsrK residues that contact with ATP (marked with A) and HPr (marked with numbers) are indicated in the bottom of the aligned sequences. The letters and numbers are colored following the characteristics of the interactions (hydrophobic, red; hydrogen bond, green; ionic interaction, blue), and the other neighboring residues remain as black.
