Abstract
Breast cancer (BC) is the leading malignancy in women worldwide, yet relatively little is known about the genes and signaling pathways involved in BC tumorigenesis and progression. The present study aimed to elucidate potential key candidate genes and pathways in BC. Five gene expression profile data sets (GSE22035, GSE3744, GSE5764, GSE21422 and GSE26910) were downloaded from the Gene Expression Omnibus (GEO) database, which included data from 113 tumorous and 38 adjacent non-tumorous tissue samples. Differentially expressed genes (DEGs) were identified using t-tests in the limma R package. These DEGs were subsequently investigated by pathway enrichment analysis and a protein-protein interaction (PPI) network was constructed. The most significant module from the PPI network was selected for pathway enrichment analysis. In total, 227 DEGs were identified, of which 82 were upregulated and 145 were downregulated. Pathway enrichment analysis results revealed that the upregulated DEGs were mainly enriched in ‘cell division’, the ‘proteinaceous extracellular matrix (ECM)’, ‘ECM structural constituents’ and ‘ECM-receptor interaction’, whereas downregulated genes were mainly enriched in ‘response to drugs’, ‘extracellular space’, ‘transcriptional activator activity’ and the ‘peroxisome proliferator-activated receptor signaling pathway’. The PPI network contained 174 nodes and 1,257 edges. DNA topoisomerase 2-a, baculoviral inhibitor of apoptosis repeat-containing protein 5, cyclin-dependent kinase 1, G2/mitotic-specific cyclin-B1 and kinetochore protein NDC80 homolog were identified as the top 5 hub genes. Furthermore, the genes in the most significant module were predominantly involved in ‘mitotic nuclear division’, ‘mid-body’, ‘protein binding’ and ‘cell cycle’. In conclusion, the DEGs, relative pathways and hub genes identified in the present study may aid in understanding of the molecular mechanisms underlying BC progression and provide potential molecular targets and biomarkers for BC.
Keywords: breast cancer, integrated bioinformatics analysis, differentially expressed genes
Introduction
Breast cancer (BC) is one of the most common types of cancer in women worldwide and its incidence is increasing, particularly in developed countries (1–3). Women are typically screened for BC with mammography and traditional tumor markers, including carcinoembryonic antigen and carcinoma antigen 15-3 (4). However, the diagnostic power of these methods is limited, due to low sensitivity and specificity (5,6). BC treatment commonly includes surgical resection and hormone therapy, radiotherapy or chemotherapy. However, BC remains highly prevalent and malignant due to recurrence and metastasis. Therefore, there is an urgent need to develop novel diagnostic strategies and therapeutic agents to improve the prognosis of patients with BC.
The molecular mechanisms of BC tumorigenesis and progression remain unclear. It is therefore critical to identify new genes and pathways that are associated with BC tumorigenesis and patient prognosis, which may not only help to elucidate the underlying molecular mechanisms involved, but also to discover novel diagnostic markers and therapeutic targets. Microarrays can rapidly detect gene expression on a global basis and are particularly useful in screening for differentially expressed genes (DEGs) (7). Gene chips are a form of microarray which allow the investigation of gene expression in a high throughput manner with high sensitivity, specificity and repeatability. A significant amount of data has been produced via the use of microarrays and the majority of such data has been uploaded and stored in public databases. Previous studies concerning BC gene expression profiling have identified hundreds of DEGs (8–10). However, the comparative analysis of DEGs across a range of independent studies may yield only a relatively limited amount of useful data with regard to carcinogenesis. The disadvantages of these single studies may be overcome by combining microarray technology and bioinformatics analysis, as this approach would make it possible to analyze the associated pathways and interaction networks associated with the identified DEGs. This information may aid in elucidating the molecular mechanisms underlying BC.
In the present study, five profiling microarray datasets were downloaded from the Gene Expression Omnibus (GEO): GSE22035, GSE3744, GSE5764, GSE21422 and GSE26910. DEGs were identified in tumor tissues relative to adjacent non-cancerous tissues in patients with BC. Additionally, gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed and protein-protein interaction (PPI) networks were constructed to identify the hub genes in BC. Collectively, the findings of the present study highlighted key genes and pathways that may contribute to the pathology of BC. These may provide a basis for the development of future diagnostic and therapeutic tools for BC.
Materials and methods
Microarray data
GSE22035 (11), GSE3744 (12), GSE5764 (13), GSE21422 (14) and GSE26910 (15) profile datasets were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) and based on a GeneChip Human Genome U133 Plus 2.0 Array platform (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The datasets contained 151 tissue samples, including 88 invasive ductal carcinoma tissues, 23 invasive lobular carcinoma tissues, five ductal carcinoma in situ tissues and 38 adjacent non-tumorous tissues.
Expression analysis of DEGs
Raw data were converted into an expression matrix, which was subsequently normalized with the robust multi-array average algorithm (16) in the affy package (version 3.4.1) in R (17). Inter-batch difference was rectified using the ComBat function in the sva R package (18). The t-test method in the limma (19) R package was subsequently used to identify DEGs between the tumor tissues and adjacent non-tumorous tissue samples. A |log2-fold change|>1 and P<0.05 were considered as the threshold values for DEG identification.
GO and pathway enrichment analysis
The Database for Annotation Visualization and Integrated Discovery (DAVID) (20) is a tool which provides a comprehensive set of functional annotation tools for researchers to investigate the biological meaning of genes. Identified DEGs were investigated further using DAVID (version 6.7), GO (21) and KEGG (22) pathway enrichment analyses. P<0.05 and gene counts of >5 were considered to indicate a statistically significant difference in the functional enrichment analysis.
Integration of the PPI network
Identified DEGs were mapped into the online Search Tool for the Retrieval of Interacting Genes (STRING; 2017 release) database (23) to evaluate the interactive relationships among the DEGs. Interactions with a combined score >0.4 were defined as statistically significant. Cytoscape software (version 3.5.1) (24) was used to visualize the integrated regulatory networks. The Cytoscape plugin Molecular Complex Detection (MCODE; version 1.31) was used to further detect deeper connected regions within the PPI network (25). According to the degree levels in the Cytoscape plugin cytoHubba (version 0.1), the top five ranked genes were defined as hub genes.
Results
The identification of DEGs in BC
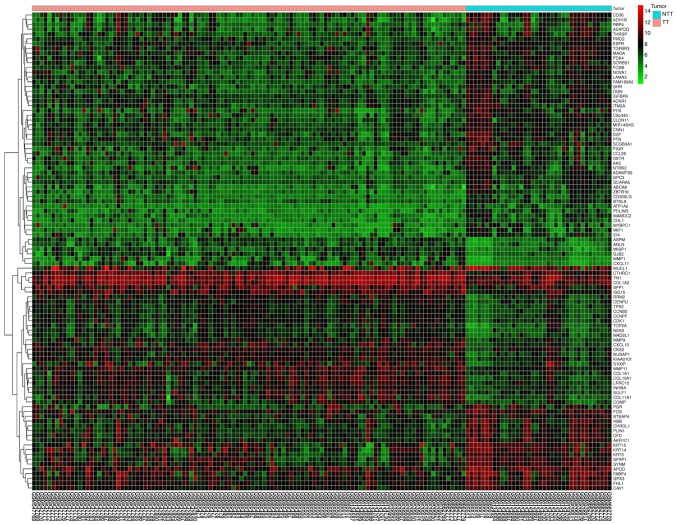
Database analysis (Table I) identified a total of 227 DEGs, including 82 upregulated genes and 145 downregulated genes. An expression heat map (Fig. 1) and a volcano plot (Fig. 2) for the identified DEGs was constructed.
Table I.
Compared with adjacent non-tumorous tissues, 227 DEGs were identified from the datasets analyzed, 82 of which were upregulated genes and 145 which were downregulated genes in breast cancer tissues.
| DEGs | Genes |
|---|---|
| Upregulated | DLGAP5, APOBEC3B, RAD51AP1, ASPN, BIRC5, LMNB1, NDC80, BGN, KIF4A, GINS1, RACGAP1, IFI6, EZH2, KIF20A, CMPK2, CCNB1, UHRF1, SQLE, VCAN, UBE2C, PTTG1, PPAPDC1A, BUB1, COL5A2, TYMS, RSAD2, FNDC1, DTL, CCNE2, BUB1B, ECT2, CXCR4, UBE2T, CDKN3, ADAMDEC1, PBK, LYZ, MELK, FAM83D, POSTN, STAT1, CXCL9, ZWINT, CEP55, PRR11, HMMR, PRC1, CEACAM6, CKS2, CDK1, CCNB2, TPX2, MMP9, COL1A2, ANLN, CENPU, MAD2L1, ISG15, CENPF, COL1A1, NEK2, CXCL11, SULF1, MMP1, GJB2, FN1, MMP11, ASPM, SPP1, INHBA, COMP, KIAA0101, WISP1, LRRC15, TOP2A, NUSAP1, RRM2, S100P, CTHRC1, CXCL10, COL10A1, COL11A1 |
| Downregulated | ADH1B, KRT14, C2orf40, DST, SFRP1, CD36, PI15, SYNM, NTRK2, ZBTB16, ABCA8, MAMDC2, OXTR, FABP4, MUCL1, PDK4, TGFBR3, KRT15, WIF1, FOSB, CFD KRT5, CHRDL1, FMO2, GPC3, APOD, ADAMTS5, PLIN1, MAOA, SCGB3A1, ID4, AK5, FHL1, CD300LG, THRSP, CLDN11, CAV1, SORBS1, LAMA3, SCARA5, FAM189A2, MIR143HG, PIGR, ACKR1, GHR, ATP1A2, RBP4, PDLIM3, CCL28, CHL1, FOS, STEAP4, BTNL9, AKR1C1, ADIPOQ, EGFR, MYBPC1, IGFBP6, ITM2A, OGN, GPX3, NOVA1, PGR, HBB, EGR1, MYH11, TF, LIFR, CXCL12, KIT, ACACB, CXCL14, SYNPO2, DMD, TFPI2, KLHL13, SRPX, SEMA3G, LEP, ENPP2, HOXA5, AKR1C3, RHOJ, ACTG2, SDPR, MIR205HG, ANXA3, LPL, ATF3, KLF4, PAMR1, TMTC1, SEMA6D, CRYAB MEOX1, HLF, ANPEP, LYVE1, ADIRF, WLS, SOCS2, ALDH1A1, AQP1, PLIN4, SPRY2, THRB, G0S2, TSHZ2, TM4SF18, ITIH5, EFEMP1, CAV2, CX3CL1, TCEAL7, EGR3, SCN4B, PPL, C16orf89, EBF1, MAOB, ANXA1, MME, ANK2, ABCA6, CXCL2, IRX1, MIR100HG, LMOD1, NDRG2, NFIB, SAMD5, AOC3, NR3C2, LHFP, COL6A6, CITED1, CDO1, BOC, TIMP4, INHBB, GSN |
DEG, differentially expressed gene.
Figure 1.
Heat map of the top 100 differentially expressed genes. A total of 82 upregulated genes and 145 downregulated genes were identified. Red indicates upregulation and green indicates downregulation. TT, tumor tissue; NTT, adjacent non-tumorous tissue.
Figure 2.
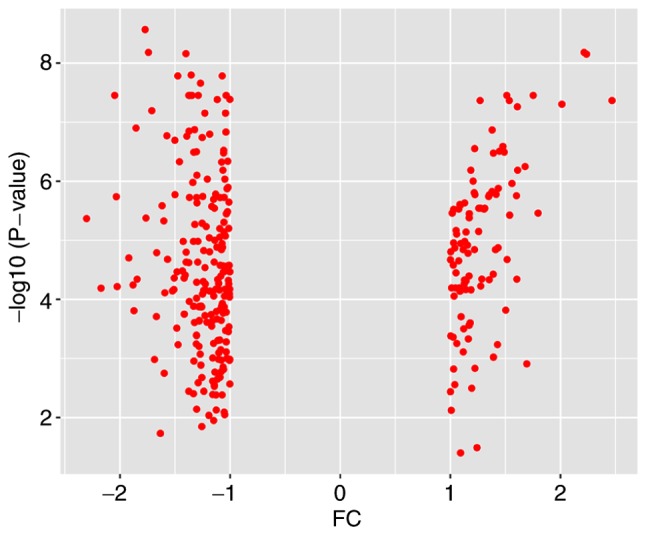
Volcano plot of the 227 identified DEGs. Red indicates DEGs with a |log2FC|>1. DEG, differentially expressed gene; FC, fold change.
Functional enrichment analysis
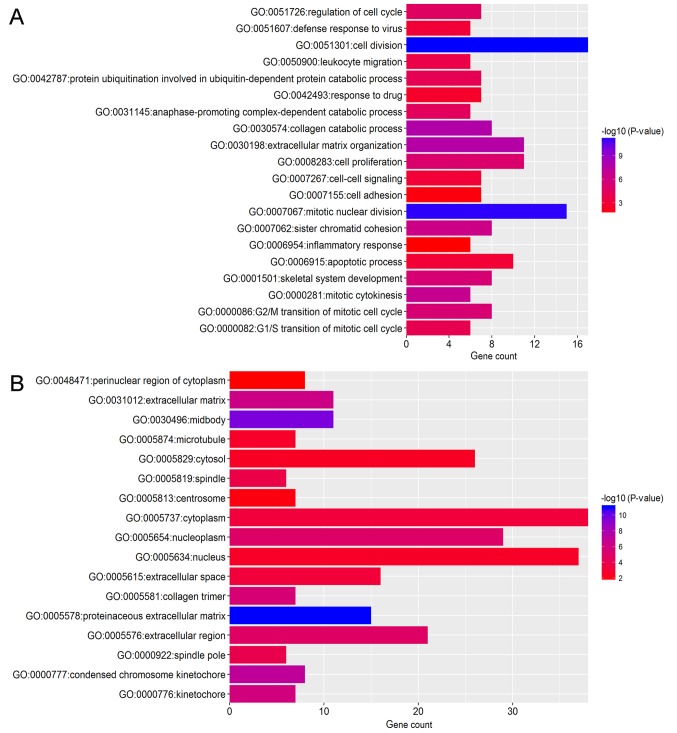
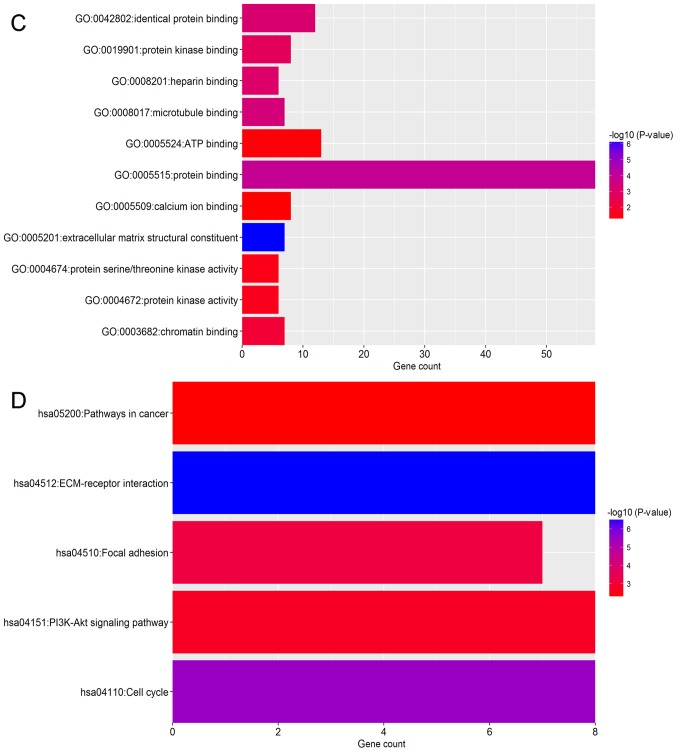
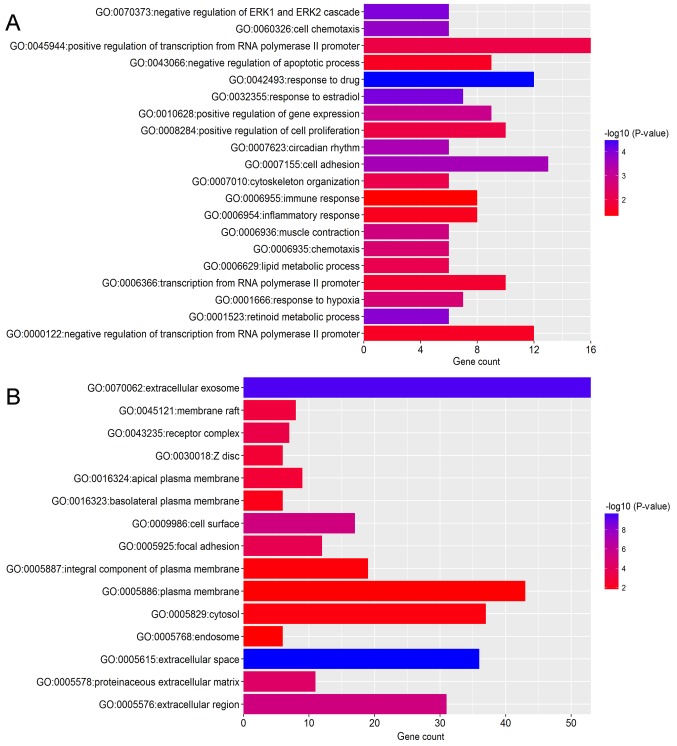
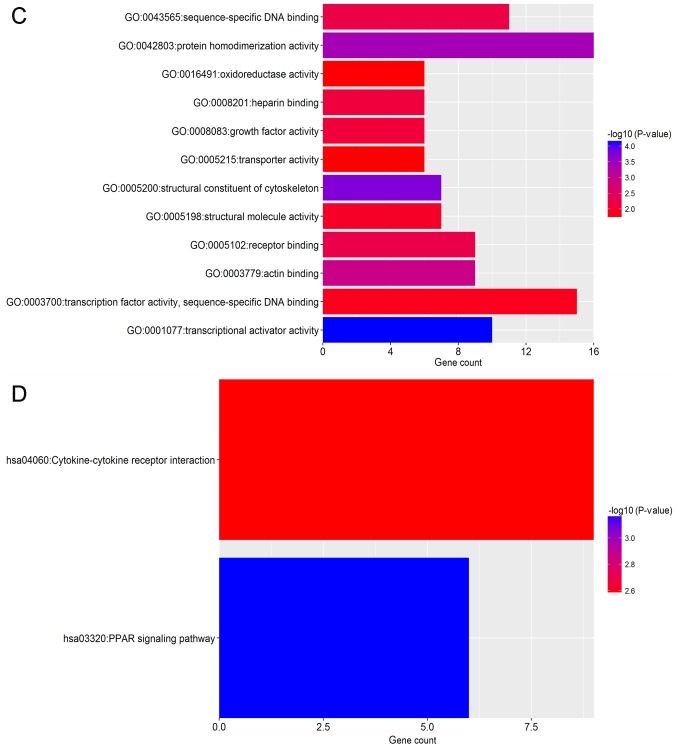
To identify the pathways which had the most significant involvement with the genes identified, upregulated (Fig. 3) and downregulated (Fig. 4) DEGs were submitted into DAVID for GO and KEGG pathway analysis. GO analysis revealed that in biological process terms, the upregulated DEGs were mainly enriched in ‘cell division’, ‘mitotic nuclear division’ and ‘collagen catabolic process’ (Fig. 3A). Downregulated DEGs were mainly enriched in ‘response to drug’, ‘response to estradiol’ and ‘negative regulation of the extracellular signal-regulated kinase (ERK)1 and ERK2 cascade’ (Fig. 4A). In cell component terms, upregulated DEGs were mainly enriched in ‘proteinaceous extracellular matrix (ECM)’, ‘mid-body’ and ‘condensed chromosome kinetochores’ (Fig. 3B), whereas downregulated DEGs were mainly enriched in ‘extracellular space’, ‘extracellular exosomes’ and the ‘extracellular region’ (Fig. 4B). In molecular function terms, upregulated DEGs were mainly enriched in ‘ECM structural constituents’, ‘protein binding’ and ‘microtubule binding’ (Fig. 3C), whereas downregulated DEGs were mainly enriched in ‘transcriptional activator activity’, ‘structural constituents of the cytoskeleton’ and ‘protein homo-dimerization activity’ (Fig. 4C).
Figure 3.
Functional enrichment analysis of upregulated DEGs in breast cancer. GO analysis revealed that DEGs were significantly enriched in (A) biological process terms, (B) cell component terms and (C) molecular function terms. (D) Significantly enriched KEGG terms obtained from KEGG analysis. DEG, differentially expressed gene; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, gene ontology.
Figure 4.
Functional enrichment analysis of downregulated DEGs in breast cancer. GO analysis revealed that DEGs were significantly enriched in (A) biological process terms, (B) cell component terms and (C) molecular function terms. (D) Significantly enriched KEGG terms obtained from KEGG analysis. DEG, differentially expressed gene; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, gene ontology.
KEGG pathway analysis demonstrated that upregulated DEGs were significantly enriched in ‘ECM-receptor interaction’, ‘cell cycle’, ‘focal adhesion’ and ‘phosphatidylinositol 3 kinase-protein kinase B (PI3K-Akt) signaling pathway’ and ‘pathways in cancer’ (Fig. 3D). Downregulated DEGs were significantly enriched in the ‘peroxisome proliferator-activated receptor (PPAR) signaling pathway’ and ‘cytokine-cytokine receptor interaction’ (Fig. 4D).
PPI network construction and module analysis
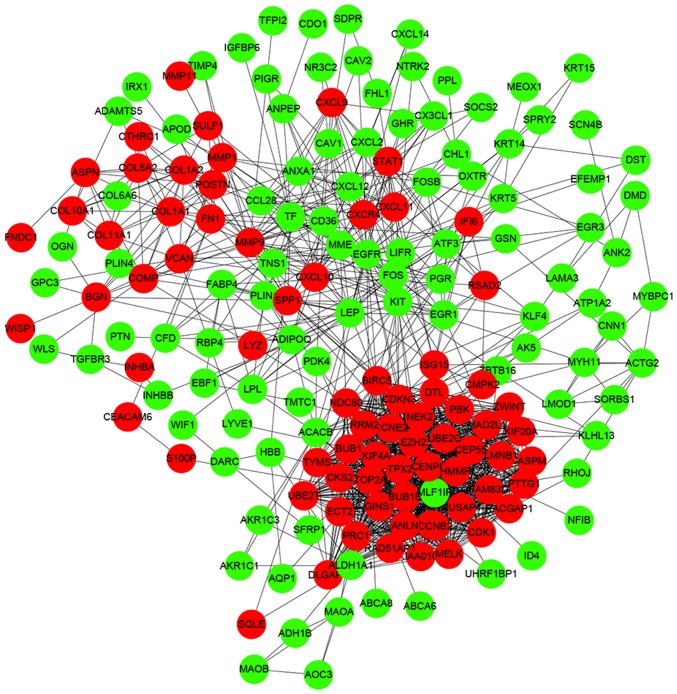
Interactions between the identified DEGs were revealed by constructing a PPI network. In total, there were 174 nodes and 1,257 edges in the network (Fig. 5). According to degree levels, the top five hub nodes were: DNA topoisomerase 2-α (TOP2A; degree, 53), baculoviral inhibitor of apoptosis repeat-containing protein 5 (BIRC5; degree, 49), cyclin-dependent kinase 1 (CDK1; degree, 45), G2/mitotic-specific cyclin-B1 (CCNB1; degree, 44) and kinetochore protein NDC80 homolog (NDC80; degree, 44). A significant module was subsequently constructed with 39 nodes and 728 edges, which gained the highest MCODE score (Fig. 6). Subsequent functional enrichment analysis revealed that the genes in this module were mainly enriched in ‘mitotic nuclear division’, ‘cell division’, ‘nucleus’, ‘cytoplasm’, ‘protein binding’, ‘ATP binding’ and ‘cell cycle’ (Table II).
Figure 5.
Protein-protein interaction network of the identified differentially expressed genes. Green indicates the downregulated genes and red indicates the upregulated genes.
Figure 6.
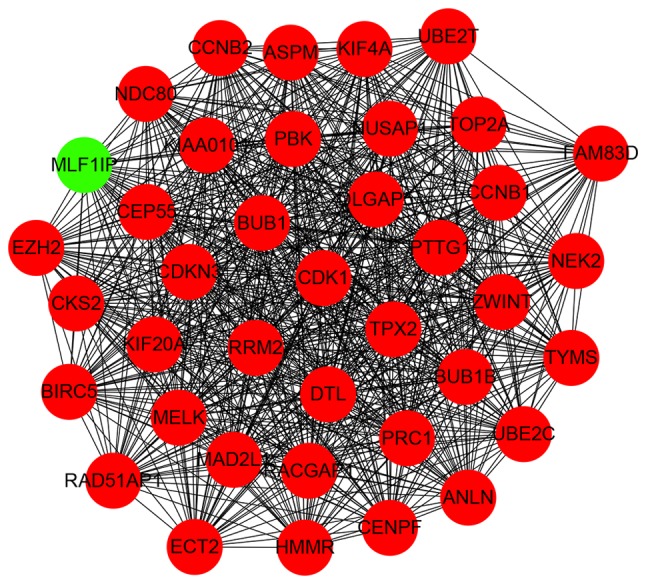
MCODE identification of the most significantly enriched module. The module with the highest MCODE score was selected from the protein-protein interaction network. Green indicates the downregulated genes and red indicates the upregulated genes.
Table II.
Pathway enrichment analysis of gene function within the identified module.
| Category | GO ID | Term | Count |
|---|---|---|---|
| BP | GO:0007067 | Mitotic nuclear division | 15 |
| BP | GO:0051301 | Cell division | 16 |
| BP | GO:0000281 | Mitotic cytokinesis | 6 |
| BP | GO:0000086 | G2/M transition of mitotic cell cycle | 8 |
| BP | GO:0008283 | Cell proliferation | 10 |
| BP | GO:0007062 | Sister chromatid cohesion | 7 |
| BP | GO:0031145 | Anaphase-promoting complex-dependent catabolic process | 6 |
| BP | GO:0042787 | Protein ubiquitination involved in ubiquitin-dependent Protein catabolic process | 6 |
| BP | GO:0006915 | Apoptotic process | 6 |
| CC | GO:0030496 | Midbody | 11 |
| CC | GO:0000776 | Kinetochore | 7 |
| CC | GO:0000777 | Condensed chromosome kinetochore | 7 |
| CC | GO:0005654 | Nucleoplasm | 21 |
| CC | GO:0005634 | Nucleus | 28 |
| CC | GO:0005829 | Cytosol | 21 |
| CC | GO:0005737 | Cytoplasm | 26 |
| CC | GO:0000922 | Spindle pole | 6 |
| CC | GO:0005819 | Spindle | 6 |
| CC | GO:0005874 | Microtubule | 7 |
| CC | GO:0005813 | Centrosome | 7 |
| MF | GO:0005515 | Protein binding | 34 |
| MF | GO:0008017 | Microtubule binding | 7 |
| MF | GO:0019901 | Protein kinase binding | 7 |
| MF | GO:0003682 | Chromatin binding | 7 |
| MF | GO:0005524 | ATP binding | 12 |
| MF | GO:0004672 | Protein kinase activity | 6 |
| MF | GO:0004674 | Protein serine/threonine kinase activity | 6 |
| MF | GO:0042803 | Protein homodimerization activity | 6 |
| KEGG | hsa04110 | Cell cycle | 7 |
GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function.
Discussion
BC is a malignant tumor that can be caused by various factors, including genetics, the endocrine system and the environment (26,27). It is critical to understand the molecular mechanisms underlying BC in order to identify and develop more effective diagnostic and therapeutic strategies. Microarray and high throughput sequencing are widely used to detect the expression levels of thousands of genes within the human genome and may aid in the identification of target genes of interest for diagnosing or treating BC (28,29).
In the present study, five gene profile datasets were obtained from GEO and bioinformatics analysis was performed, resulting in identification of 227 genes which were differentially expressed between BC and normal controls. Functional enrichment analysis revealed that upregulated genes were mainly enriched in ‘cell division’, ‘proteinaceous ECM’, ‘ECM structural constituents’ and ‘ECM-receptor interaction’, whereas downregulated genes were mainly enriched in ‘response to drugs’, ‘extracellular space’, ‘transcriptional activator activity’ and the ‘PPAR signaling pathway’. A PPI network was constructed for the identified DEGs and key genes were defined by the degree rank. The most significant module was subsequently extracted from the PPI network.
Previous research involving multiple cohort studies tend to have a lower false-positive and false-negative rate than single cohort studies (30). However, multiple microarrays from different platforms may mask and confound true biological differences because of the batch effects (31). In order to increase the credibility of DEG identification, five microarray datasets from the same platform were selected and a ComBat function was used to eliminate batch effect in the present study. A total of 227 genes were identified which were differentially expressed between tumor tissues and the adjacent non-tumorous tissues, including 82 upregulated genes and 145 downregulated genes. It has been demonstrated that there is a co-expression association between a group of genes with similar expression profiles and these often participate in parallel biological processes (32). Therefore, it is necessary to perform functional enrichment analysis in order to understand the interactions between DEGs and the associated biological processes.
The upregulated genes identified in the present study were mainly enriched in pathways of ‘cell division’, ‘proteinaceous ECM’, ‘ECM structural constituents’ and ‘ECM-receptor interaction’. This is consistent with the fact that the ECM is an important component in the mammary gland microenvironment and that ECM proteins have been demonstrated to accelerate BC tumor progression and metastasis (33–36). Furthermore, previous studies have indicated that the gene expression signatures of the BC stroma, which includes ECM proteins, can better predict patient outcome than the tumorous epithelium (15,37,38).
The downregulated DEGs identified in the present study were associated with ‘response to drugs’, ‘extracellular space’, ‘transcriptional activator activity’ and the ‘PPAR signaling pathway’. PPARs are members of the nuclear hormone receptor superfamily and function in proliferation, differentiation, inflammation and glucose and lipid balance (39). Evidence suggests that PPARg ligands may be regarded as antitumor factors in humans due to their involvement in apoptosis and cell growth inhibition in several malignant tumor cell types, including colon adenocarcinoma, hepatocellular carcinoma and breast cancer (40). PPARg has also emerged as a potential target for cancer therapy as it has high tumor specificity (41). It has been demonstrated that PPARg inhibits the invasion and metastasis of human BC cells (42). Furthermore, evidence has indicated that PPARg expression and signaling in mammary secretory epithelial cells has a protective role against breast tumorigenesis (43).
By constructing a PPI network, TOP2A, BIRC5, CDK1, CCNB1 and NDC80 were identified to have higher degrees of connectivity within the network and were therefore classified as the hub genes in the present study. Additionally, module analysis of the PPI network revealed that BC development was associated with ‘mitotic nuclear division’, ‘mid-body’, ‘protein binding’ and ‘cell cycle’.
TOP2A is an enzyme that generates transient double-stranded breaks in the topological structure of DNA (44). In a previous report, Milde-Langosch et al (45) reported that TOP2A expression may be regarded as an indicator of susceptibility to anthracycline neoadjuvant therapy in BC. Additionally, Şahin et al (46) demonstrated that the overexpression of TOP2A is associated with poor prognosis in patients with BC.
BIRC5 is a member of the inhibitor of apoptosis gene family and is located on chromosome 17q25 (47). BIRC5 is involved in cell cycle checkpoint progression and is overexpressed in breast carcinomas; the degree of overexpression correlates with poor patient outcome (48,49). Consequently, the BIRC5 gene is a potential marker for the detection and prognosis of cancer at an early age (50).
CDK1 is a conserved serine/threonine kinase that controls cell cycle progression and is essential for driving the cell cycle (51). A previous study reported that CDK1 is overexpressed in BC (52). Additionally, CDK1 degradation may be mediated by sequestosome-1-histone deacetylase 6-dependent autophagy and the aggresome pathway in BC (53). Consequently, it has been reported that the levels of CDK1 clearance may be a predictive biomarker for the efficacy of BC chemotherapy (53).
CCNB1 is a highly conserved member of the cyclin family that is expressed in almost all tissues of the human body (54). CCNB1 is a key initiator of mitosis through regulation of CDK1, which is responsible for initiating progression from the G2 phase to mitosis (55). Other research has revealed that CCNB1 is expressed in many types of cancer, suggesting that it may also function in cancer transformation and progression (56). Ding et al (57) demonstrated that CCNB1 expression may be used to monitor hormone therapy efficacy and this may aid in the development of personalized therapies for patients with estrogen receptor+ BC.
NDC80 is a nuclear protein rich in coiled-coil motifs which was first discovered by Durfee et al (58) by combining the C-terminus of the retinoblastoma protein and using yeast two-hybrid technology to screen a B-lymphocyte complementary DNA library. NDC80 is a kinetochore outer layer component and spindle checkpoint regulator that is involved in chromosome segregation via overactivation of the mitotic checkpoint (59). Furthermore, it has been reported that NDC80 overexpression may participate in tumor formation by activating the mitotic checkpoint and is associated with poor clinical prognosis in patients with BC (60,61).
In conclusion, the present study identified candidate genes and pathways which may be involved in BC progression through the integrated analysis of multiple cohort profile datasets. These results may contribute to a better understanding of the molecular mechanisms which underlie BC and provide a series of potential biomarkers. However, further experiments are required to verify the findings of the present study. Additionally, the majority of included studies focused on how a single key gene and pathway contribute to the development of tumor in breast cancer, with limited research concerning the interaction of multi-genes and multi-pathways. Therefore, further experiments with additional patient cohorts are also required to confirm the results of this study. In vivo and in vitro investigation of gene and pathway interaction is essential to delineate the specific roles of the identified genes, which may help to confirm gene functions and reveal the mechanisms underlying BC.
Acknowledgements
The authors thank Kaijiong Zhang for his technical guidance.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CL and YW conceived and designed the study. QH and YZ collected the data. YW and YZ analyzed the database, prepared the diagrams and wrote the paper. CL and QH reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Fan L, Strasser-weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 3.Elfar GA, Ebrahim MA, Elsherbiny NM, Eissa LA. Validity of osteoprotegerin and receptor activator of NF-κB ligand for the detection of bone metastasis in breast cancer. Oncol Res. 2017;25:641–650. doi: 10.3727/096504016X14768398678750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uehara M, Kinoshita T, Hojo T, Akashi-Tanaka S, Iwamoto E, Fukutomi T. Long-term prognostic study of carcinoembryonic antigen (CEA) and carbohydrate antigen 15-3 (CA 15-3) in breast cancer. Int J Clin Oncol. 2008;13:447–451. doi: 10.1007/s10147-008-0773-3. [DOI] [PubMed] [Google Scholar]
- 5.Duffy MJ, Evoy D, McDermott EW. CA 15-3: Uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411:1869–1874. doi: 10.1016/j.cca.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 6.O'Donoghue C, Eklund M, Ozanne EM, Esserman LJ. Aggregate cost of mammography screening in the United States: Comparison of current practice and advocated guidelines. Ann Intern Med. 2014;160:145. doi: 10.7326/M13-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Ellis MJ. Microarray data analysis in neoadjuvant biomarker studies in estrogen receptor-positive breast cancer. Breast Cancer Res. 2010;12:112. doi: 10.1186/bcr2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Li Z, Yang Y, Xiang T, Song W, Liu S. Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer. Cancer Biol Ther. 2015;16:856–865. doi: 10.1080/15384047.2015.1040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makoukji J, Makhoul NJ, Khalil M, El-Sitt S, Aldin ES, Jabbour M, Boulos F, Gadaleta E, Sangaralingam A, Chelala C, et al. Gene expression profiling of breast cancer in Lebanese women. Sci Rep. 2016;6:36639. doi: 10.1038/srep36639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cizkova M, Cizeron-Clairac G, Vacher S, Susini A, Andrieu C, Lidereau R, Bièche I. Gene expression profiling reveals new aspects of PIK3CA mutation in ERalpha-positive breast cancer: Major implication of the Wnt signaling pathway. PLoS One. 2010;5:e15647. doi: 10.1371/journal.pone.0015647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, Klein J, Fridman E, Skarda J, Srovnal J, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Planche A, Bacac M, Provero P, Fusco C, Delorenzi M, Stehle JC, Stamenkovic I. Identification of prognostic molecular features in the reactive stroma of human breast and prostate cancer. PLoS One. 2011;6:e18640. doi: 10.1371/journal.pone.0018640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 17.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, Liu C. Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLoS One. 2011;6:e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao X, Sherman BT, da Huang W, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gene Ontology Consortium: The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray JM, Rasanayagam S, Engel C, Rizzo J. State of the evidence 2017: An update on the connection between breast cancer and the environment. Environ Health. 2017;16:94. doi: 10.1186/s12940-017-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamdi Y, Soucy P, Adoue V, Michailidou K, Canisius S, Lemaçon A, Droit A, Andrulis IL, Anton-Culver H, Arndt V, et al. Association of breast cancer risk with genetic variants showing differential allelic expression: Identification of a novel breast cancer susceptibility locus at 4q21. Oncotarget. 2016;7:80140–80163. doi: 10.18632/oncotarget.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng T, Wang A, Hu D, Wang Y. Molecular mechanisms of breast cancer metastasis by gene expression profile analysis. Mol Med Rep. 2017;16:4671–4677. doi: 10.3892/mmr.2017.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda K, Horie-Inoue K, Inoue S. Identification of estrogen-responsive genes based on the DNA binding properties of estrogen receptors using high-throughput sequencing technology. Acta Pharmacol Sin. 2015;36:24–31. doi: 10.1038/aps.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of P-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- 31.Kupfer P, Guthke R, Pohlers D, Huber R, Koczan D, Kinne RW. Batch correction of microarray data substantially improves the identification of genes differentially expressed in rheumatoid arthritis and osteoarthritis. BMC Med Genomics. 2012;5:23. doi: 10.1186/1755-8794-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33:111. doi: 10.1007/s12032-016-0829-6. [DOI] [PubMed] [Google Scholar]
- 33.Lyons TR, O'Brien J, Borges VF, Conklin MW, Keely PJ, Eliceiri KW, Marusyk A, Tan AC, Schedin P. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat Med. 2011;17:1109–1115. doi: 10.1038/nm.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG, Keely PJ. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox TR, Bird D, Baker AM, Barker HE, Ho MW, Lang G, Erler JT. LOX-mediated collagen crosslinking is responsible for fibrosis-enhanced metastasis. Cancer Res. 2013;73:1721–1732. doi: 10.1158/0008-5472.CAN-12-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chia J, Kusuma N, Anderson R, Parker B, Bidwell B, Zamurs L, Nice E, Pouliot N. Evidence for a role of tumor-derived laminin-511 in the metastatic progression of breast cancer. Am J Pathol. 2007;170:2135–2148. doi: 10.2353/ajpath.2007.060709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sørlie T, Dai H, He YD, van't Veer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 39.Peters JM, Shah YM, Gonzalez FJ. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12:181–195. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boitier E, Gautier JC, Roberts R. Advances in understanding the regulation of apoptosis and mitosis by peroxisome-proliferator activated receptors in pre-clinical models: Relevance for human health and disease. Comp Hepatol. 2003;2:3. doi: 10.1186/1476-5926-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robbins GT, Nie D. PPAR gamma, bioactive lipids, and cancer progression. Front Biosci (Landmark Ed) 2012;17:1816–1834. doi: 10.2741/4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, Zang C, Fenner MH, Possinger K, Elstner E. PPARgamma ligands and ATRA inhibit the invasion of human breast cancer cells in vitro. Breast Cancer Res Treat. 2003;79:63–74. doi: 10.1023/A:1023366117157. [DOI] [PubMed] [Google Scholar]
- 43.Apostoli AJ, Skelhorne-Gross GE, Rubino RE, Peterson NT, Di Lena MA, Schneider MM, SenGupta SK, Nicol CJ. Loss of PPARγ expression in mammary secretory epithelial cells creates a pro-breast tumorigenic environment. Int J Cancer. 2014;134:1055–1066. doi: 10.1002/ijc.28432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan J, Huang HY, Lee SW, Chen TJ, Tai HC, Hsu HP, Chang KY, Li CF. TOP2A overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biol. 2014;35:179–187. doi: 10.1007/s13277-013-1022-6. [DOI] [PubMed] [Google Scholar]
- 45.Milde-Langosch K, Karn T, Müller V, Witzel I, Rody A, Schmidt M, Wirtz RM. Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast Cancer Res Treat. 2013;137:57–67. doi: 10.1007/s10549-012-2296-x. [DOI] [PubMed] [Google Scholar]
- 46.Şahin S, Işık Gönül İ, Çakır A, Seçkin S, Uluoğlu Ö. Clinicopathological significance of the proliferation markers Ki67, RacGAP1, and Topoisomerase 2 alpha in breast cancer. Int J Surg Pathol. 2016;24:607–613. doi: 10.1177/1066896916653211. [DOI] [PubMed] [Google Scholar]
- 47.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin cancer Res. 2000;6:127–134. [PubMed] [Google Scholar]
- 49.Kappler M, Kotzsch M, Bartel F, Füssel S, Lautenschläger C, Schmidt U, Würl P, Bache M, Schmidt H, Taubert H, Meye A. Elevated expression level of survivin protein in soft-tissue sarcomas is a strong independent predictor of survival. Clin Cancer Res. 2003;9:1098–1104. [PubMed] [Google Scholar]
- 50.Ghaffari K, Hashemi M, Ebrahimi E, Shirkoohi R. BIRC5 genomic copy number variation in early-onset breast cancer. Iran Biomed J. 2016;20:241–245. doi: 10.7508/ibj.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 52.Kourea HP, Koutras AK, Scopa CD, Marangos MN, Tzoracoeleftherakis E, Koukouras D, Kalofonos HP. Expression of the cell cycle regulatory proteins p34cdc2, p21waf1, and p53 in node negative invasive ductal breast carcinoma. Mol Pathol. 2003;56:328–335. doi: 10.1136/mp.56.6.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galindo-Moreno M, Giráldez S, Sáez C, Japón MÁ, Tortolero M, Romero F. Both p62/SQSTM1-HDAC6-dependent autophagy and the aggresome pathway mediate CDK1 degradation in human breast cancer. Sci Rep. 2017;7:10078. doi: 10.1038/s41598-017-10506-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyazaki T, Arai S. Two distinct controls of mitotic cdk1/cyclin B1 activity requisite for cell growth prior to cell division. Cell Cycle. 2007;6:1419–1425. doi: 10.4161/cc.6.12.4409. [DOI] [PubMed] [Google Scholar]
- 55.Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- 56.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 57.Ding K, Li W, Zou Z, Zou X, Wang C. CCNB1 is a prognostic biomarker for ER+ breast cancer. Med Hypotheses. 2014;83:359–364. doi: 10.1016/j.mehy.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 58.Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 59.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos RG, Maiolica A, Polka J, De Luca JG, De WP, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diaz-Rodríguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.