Abstract
Magnetic Resonance Fingerprinting (MRF) is a new approach to quantitative magnetic resonance imaging that allows simultaneous measurement of multiple tissue properties in a single, time-efficient acquisition. The ability to reproducibly and quantitatively measure tissue properties could enable more objective tissue diagnosis, comparisons of scans acquired at different locations and time points, longitudinal follow-up of individual patients and development of imaging biomarkers. This review provides a general overview of MRF technology, current preclinical and clinical applications and potential future directions. MRF has been initially evaluated in brain, prostate, liver, cardiac, musculoskeletal imaging, and measurement of perfusion and microvascular properties through MR vascular fingerprinting.
Introduction
Magnetic Resonance Imaging (MRI) is a versatile imaging technique for producing exquisite anatomical images. As compared to other cross-sectional imaging modalities, MRI provides superior soft-tissue contrast[1] and has no ionizing radiation exposure. Present-day MR scanning focuses on changing various MR system parameters such as echo time (TE), repetition time (TR) and flip angle (FA) in a systematic manner to produce images generated that are said to be qualitatively “weighted,” most often by the T1 and T2 of the tissues. Current MR descriptions such as “hyperintense” and “hypointense” are relative descriptions and do not reflect absolute property values[2]. MRI also allows measurement of various tissue properties such as longitudinal relaxation time (T1), transverse relaxation time (T2), proton density (M0), diffusion and perfusion, but these properties, particularly T1 and T2, are not typically quantitatively measured in practice.
The last few years have seen an increasing emphasis on rapid and quantitative imaging. Quantitative MR imaging can provide data that can be used as imaging biomarkers for better characterization of tissue pathology, prognostication, follow-up, patient-specific management, and therapy design[3]. While diffusion and sometimes perfusion mapping have been accepted into clinical MRI protocols to provide a modicum of quantitative information, conventional T1 and T2 mapping techniques are limited by their time inefficiency. Conventional T1 and T2 mapping methods measure tissue properties by measuring signal changes obtained by varying a single acquisition parameter, keeping all the others constant. These approaches are relatively time-consuming and typically measure only one tissue property at a time [4,5].
A novel approach to quantitative MRI was recently introduced, called MR Fingerprinting (MRF) [2]. This technique allows simultaneous efficient measurements of multiple tissue properties with one acquisition [2]. This review provides a general overview of MRF technology, current preclinical and clinical applications and potential future directions.
MRF Description
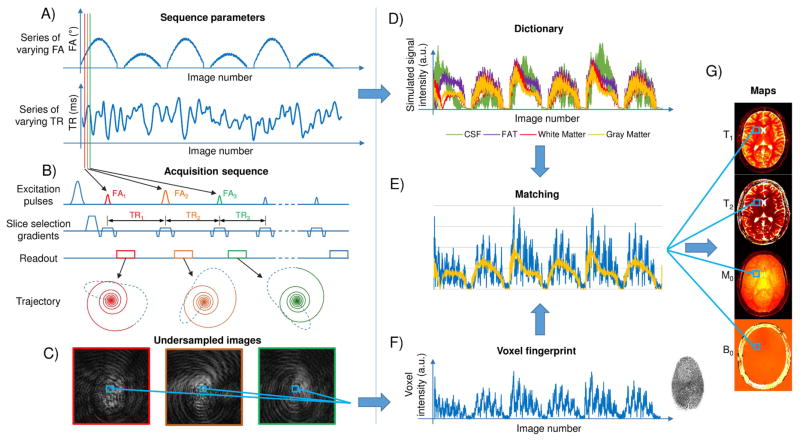
MRF can be described as a three-step process comprising of data acquisition, pattern matching and tissue property visualization. The data acquisition involves deliberately varying MR system settings and parameters, i.e. the MRF pulse-sequence, in a pseudorandom manner in order to generate unique signal evolutions, or “fingerprints”, for each combination of the tissue properties of interest. The fingerprints from individual voxels are compared with a collection of simulated fingerprints contained in a dictionary generated for that MRF sequence. The best match for the voxel fingerprint is selected from the dictionary through a pattern matching process. Once there is a pattern match, the combination of tissue properties that were used to generate the simulated fingerprint are identified as the underlying tissue properties in that voxel and these tissue properties are depicted as pixel-wise maps that are perfectly registered to one another, thereby providing quantitative and anatomic information[4] (Figure 1). Each of these steps is discussed in more detail below. While the original MRF description focused on measuring T1, T2, static magnetic field (B0) inhomogeneity or off-resonance frequency and proton density M0[2], recent work has shown the feasibility to measure other properties such as radio frequency (RF) transmit field inhomogeneity (B1)[6,7], T2*[8], perfusion[9] and microvascular properties[10–12].
Figure 1. MRF overview.
The flowchart shows an overview of the MRF framework as used for MR-True Fast Imaging with Steady State Precession (TruFISP) acquisition. (A) shows an example of variable flip angles (FA) and time of repetition (TR) used for this acquisition. (B) Sequence diagram showing the excitation pulses, slice selection gradients, readout and k-space trajectory for each TR (C) shows three undersampled images acquired in different TR. (D) shows examples of four “dictionary” entries representing four main tissues; cerebrospinal fluid (CSF) (T1= 5000ms, T2 = 500 ms) fat (T1= 400ms, T2 = 53 ms), white matter (T1 = 850 ms, T2 =50 ms) and gray matter (T1 = 1300 ms, T2 = 85 ms). (E) shows pattern matching of the voxel fingerprint with the closest entry in the dictionary, which allows to retrieve the tissue features represented by that voxel. (F) shows intensity variation of a voxel across the undersampled images. (G) shows parameter maps obtained by repeating the matching process for each voxel.
Data Acquisition
In MRF, there is a fundamental difference in the way data are acquired, as compared to conventional MRI. Instead of repeating the same acquisition parameters over time in a particular sequence until all the data in k-space have been obtained and used to reconstruct images with weighting by a particular property; in MRF the acquisition parameters such as the radiofrequency excitation angle (FA) and phase, repetition time, and k-space sampling trajectory, are varied throughout the acquisition, which when implemented properly can generate a unique signal timecourse for each tissue. Proper implementation of the sequence design is crucial for obtaining useful information and determines the relevant combination of tissue properties that can be measured, how time-efficient, accurate, precise and clinically useful that MRF sequence is.
Reduction of acquisition time is important for volumetric coverage and coverage of large body regions. As originally described, MRF acquisitions were already extremely undersampled (only 1/48th of full image data set was acquired for each time point) [2]. This undersampling results in severe artifacts in the image from each individual time point. However, a philosophical decision is made in MRF: Unlike traditional mapping techniques, the focus is solely on generating quantitative maps of interest, and the artifacts in each individual image are not of concern as long as they do not compromise the matching process. Thus high quality individual time point images are explicitly not sought. Despite the significant undersampling, the signal evolution obtained from all the undersampled data can still be matched to the best corresponding MRF dictionary entry and the resulting quantitative maps have been shown to be highly accurate[2] and repeatable (Coefficient of variation of less than 5% for T1 and T2 values)[13]. It is therefore possible to “see through” image artifacts arising from the undersampling during the dictionary-matching step, as the errors are not coherent spatially or temporally [2]. Thus, spatio-temporal incoherence of the undersampling artifacts is a primary consideration for design of the sampling strategy. For data sampling, spiral[2,14–17] or radial[7,18] trajectories are often used because of higher spatial incoherence and sampling efficiency. However, echo-planar imaging (EPI)[8,9] and Cartesian trajectories[6,10,19–21] have also been shown to be useful in MRF framework. Depending on the sequence type, the variation in acquisition parameters and the application, the trajectory re-ordering can be sequential, uniformly rotated or random. For example, Ma, et al[2] used a sequential reordering strategy with spiral trajectory for balanced steady state free precession (bSSFP) based MRF for brain imaging. Hamilton, et al[15] used a golden angle based rotational reordering strategy with spiral trajectory for steady state free precession (SSFP) based MRF for cardiac imaging. Cloos, et al[7] used a uniform rotational reordering strategy with radial trajectory for Plug-n-Play based MRF (PnP-MRF) for musculoskeletal imaging.
MRF provides a framework in which any sequence structure can be theoretically adopted to obtain relevant tissue properties[2,4]. The original MRF description was based on inversion recovery prepared balanced steady state free precession (IR-bSSFP) because this sequence is sensitive to T1, T2 and static field (B0) inhomogeneity and because the behavior of spins in this sequence has been extensively studied[22–24]. Subsequent descriptions have adapted more sequences to MRF, each in order to overcome certain limitations of other sequences, confer additional advantages or measure additional tissue properties. For example, Jiang, et al[14], utilized SSFP sequence for MRF which is insensitive to B0 inhomogeneity, eliminates banding artifacts seen with bSSFP and can be readily adapted for body applications. Ässlander, et al[18] adopted a pseudo steady-state free precession (pSSFP) sequence to improve the spin-echo like signal properties of a bSSFP based MRF acquisition and thereby reduce the effects of B0 inhomogeneity on estimated tissue properties within a limited range of B0. Reiger, et al [8]incorporated an EPI based data acquisition approach into the MRF framework and a variation in TE for simultaneously estimating T1 and T2*. EPI-MRF relaxometry values were in good agreement with gold standard values with a small coefficient of variation (T1: 4 ± 2%, T2*: 4 ± 9%)[8]. On the other hand, Su, et al[9] integrated arterial spin labeling (ASL) concept into MRF framework for estimation of perfusion related tissue properties.
Apart from modifying various sequences to MRF framework, work has also been directed to improve MR safety[25], patient comfort[26] and decrease scan time[27–32] during MRF. For example, Jiang et al[25], described a modification called quick echo splitting NMR (QUEST) based MRF sequence which reduces the number of radiofrequency (RF) pulses used during the acquisition and thereby decreases the RF energy deposition (specific absorption rate) into subjects. Taking advantage of the freedom in sequence design in MRF that lifts the constraint of preset gradient patterns, rather allowing them to be varied as desired, Ma et al[26], described music based-MRF in which the waveforms of music audio files were used to design the timing of acquisition and the gradients, changing the noise produced during MR scanning to a selected music file, which can in turn reduce patient anxiety. Reduction of scan times has been possible using simultaneous multislice imaging techniques[29,28,32] and mathematically advanced reconstruction methods[27,30–39].
Pattern matching and Tissue property visualization
Pattern matching involves matching the patterns of signal evolutions generated for individual tissue voxels, against the best corresponding entry in the overall dictionary of possible signal evolutions generated for that sequence. For every MRF sequence, the dictionary of signal evolutions can be generated on a computer using mathematical algorithms to predict spin behavior and signal evolution during that acquisition. In the original MRF sequence, Bloch Equations simulations[40] were utilized to predict spin behavior. The extended phase graph formalism[41] has also been utilized in later work on MRF to predict spin behavior. Work on MR vascular fingerprinting (MRvF)[10] and MRF-Arterial Spin labeling (ASL) perfusion [9] have used more complex models to generate database of fingerprints for pattern matching. Pattern matching provides a certain degree of tolerance to errors as long as these errors are spatially and temporally incoherent in such a way that it is possible for the pattern matching algorithm to select the correct match from the dictionary despite the individual errors or noisiness in the timecourse [2].
For the template matching process utilized in the original MRF acquisition, the vector-dot product of the acquired signal with each simulated fingerprint signal was calculated. The dictionary entry with the highest dot product was considered to be the best match and the T1, T2 and B0 values utilized to construct that entry were assigned to that voxel. The M0 value was computed as the multiplicative factor between the acquired and simulated fingerprints. This process was time efficient (requiring 160 seconds for a 2D brain acquisition on a standard desktop computer), accurate (good correlation with phantom values), precise and insensitive to motion artifacts[2].
Depending on the organ being evaluated or the physiological properties being measured through the MRF acquisition, the collection of fingerprints may either be generated only once for each sequence and applied to all patients[2,14] or may have to be generated individually for each patient[10,15]. For example in cardiac MRF as described by Hamilton, et al[15], the simulation was repeated individually for each new scan to accommodate the differences in heart rate for each person. However, this process took only 12 seconds, so there was no major time-penalty. Similarly in MRvF, the model needs to be simulated individually for each patient to account for the difference in contrast agent dosage according to patient body weight[10].
Since pattern matching and visualization needs to be fast, robust and accurate, efforts have also been spent to further speed up the matching process by compressing the whole process in the time dimension[42] or using a fast group matching algorithm[43], thereby improving overall speed of map generation. Compression methods have yielded a time-reduction factor of 3–5 times with less than 2% decrease in the accuracy of tissue property estimation[42,43].
Current MRF applications
MRF has been validated with phantom studies as well as in normal volunteers and patients. The T1 and T2 values obtained with MRF studies have shown good correlation with conventional T1 and T2 mapping methods and those published in literature[14,16,44–46]. The utility of MRF has also been demonstrated in preclinical and animal studies[12,21]. Clinical applications of MRF till date have been greatly focused on brain and prostate and shown promise in abdomen, musculoskeletal and cardiac applications.
Brain Relaxometry
The original MRF description showed good literature-correlation for T1 and T2 values of grey and white matter while there was a slight mismatch for T2 values of CSF[2]. This mismatch was attributed to the through-plane motion of CSF which was not considered during the initial dictionary stimulation[2]. Badve, et al[47] used the same MRF framework to simultaneously estimate T1 and T2 relaxometry values for different brain regions in 56 normal healthy volunteers. In this study, the acquisition time was 31 seconds per slice for a resolution of 1.2 × 1.2 × 5 mm3[47]. MRF-derived relaxometry provided simultaneous and fast estimation of T1 and T2 values as compared to conventional mapping methods and demonstrated regional differences and correlation with age and gender[47].
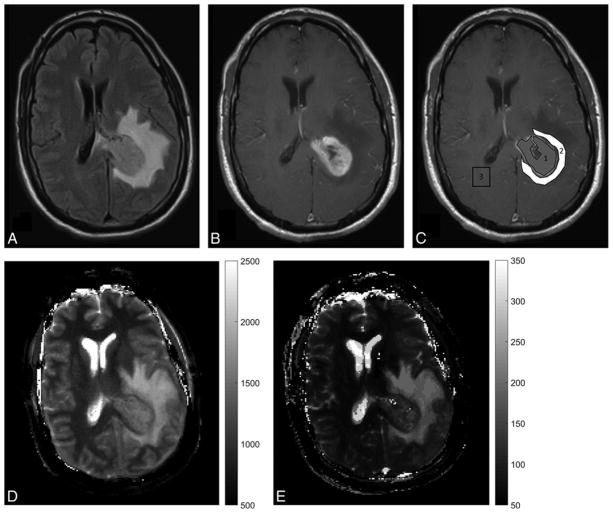
MRF has also been used to characterize and differentiate between various intra-axial brain tumors. In a study on 31 patients with intra-axial brain lesions, MRF-derived T1 and T2 values were used to differentiate gliomas and metastases[48]. The mean T2 values were significantly different for lower grade gliomas as compared to metastases (mean, 172 ± 53 ms, and 105 ± 27 ms, respectively; P = .004) while the mean T1 of peritumoral white matter surrounding lower grade gliomas differed from peritumoral white matter around glioblastomas (mean, 1066 ± 218 ms, and 1578 ± 331 ms, respectively; P = .004.) Mean T2 of solid tumors offered the best separation between glioblastomas and metastases with an area under the curve (AUC) of 0.86 (95% CI, 0.69–1.00; P < .0001)[48](Figure 2). Ongoing work on MRF in brain tumors is directed towards using a volumetric 3D MRF sequence for improved lesion visualization[49,50], assessment of treatment response, identifying radiation necrosis and tumor recurrence.
Figure 2. MRF in brain tumor.
(A, B) Conventional MR images of a patient with glioblastoma multiforme (grade 4 brain tumor) show an enhancing tumor with peritumoral white matter edema on the left side. (C) show ROIs with grey ROI segmenting the tumor and white ROI segmenting peritumoral white matter edema. (D, E) show MRF-derived quantitative T1 map T2 maps respectively. The mean T1 and T2 values for tumor were 1408 ms ± 86 ms and 112 ± 25.4 ms respectively while the mean T1 and T2 values for the peritumoral white matter were 1614 ± 101 ms and 140 ± 15.7 ms respectively.
Reprinted with permission from American Society of Neuroradiology from Figure 1, Badve C, Yu A, Dastmalchian S, Rogers M, Ma D, Jiang Y, Margevicius S, Pahwa S, Lu Z, Schluchter M, et al. MR Fingerprinting of Adult Brain Tumors: Initial Experience. AJNR Am. J. Neuroradiol. 2016, doi:10.3174/ajnr.A5035[38].
Prostate
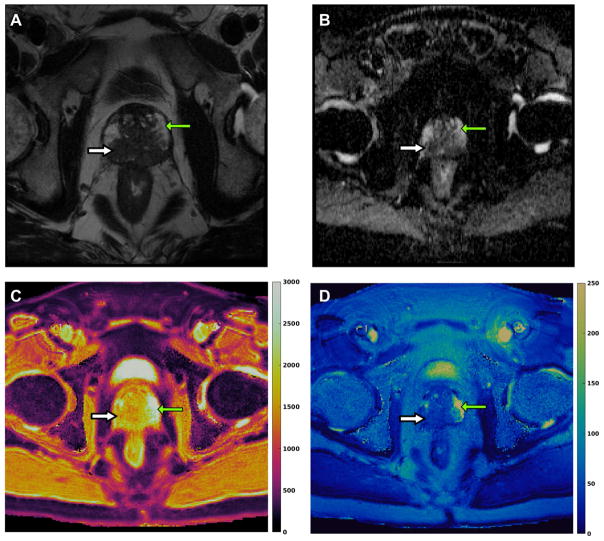
In recently published work on prostate, the feasibility of using a combined MRF –SSFP and apparent diffusion coefficient (ADC) mapping exam was evaluated for separating cancer from normal prostate in the peripheral zone of prostate[51]. MRF-SSFP sequence was added to a conventional prostate MRI protocol and regions of interest (ROIs) were drawn on MRF T1 and T2 maps and DWI images using the conventional T2 weighted (T2w) image as a reference. The reported time for MRF acquisition was 39 seconds per slice for a resolution of 1 × 1 × 5 mm3[51]. The mean T1, T2, and apparent diffusion coefficient (ADC) values from cancer (mean, 1628 ± 344 msec, 73 ± 27 msec, and 0.773 × 10−3 ± 0.331 × 10−3 mm2/sec, respectively) were significantly lower than those from normal peripheral zone (mean, 2247 ± 450 msec, 169 ± 61 msec, and 1.711 × 10−3 ± 0.269 × 10−3 mm2/sec) (P < .0001 for each) and together produced the best separation between these groups (AUC = 0.99). Importantly, ADC and T2 together also produced an AUC of 0.83 for separating high- or intermediate-grade tumors from low-grade cancers[51](Figure 3).
Figure 3. MRF in prostate cancer.
(A) T2 weighted axial image of a patient with a high-grade prostate cancer (Gleason score 9) shows a large hypointense lesion involving peripheral zone (white thick arrow) while the normal peripheral zone is hyperintense (green thin arrow). (B) Apparent diffusion coefficient (ADC) map shows diffusion restriction (dark on ADC map) in prostate cancer (white thick arrow) while normal peripheral zone shows no diffusion restriction (green thin arrow). (C, D) show MRF-derived quantitative T1 and T2 color maps respectively. The mean T1 and T2 values for prostate cancer were 1533 ± 175 ms and 36.8 ± 6.7 ms respectively while the mean T1 and T2 values for the normal peripheral zone were 2920 ± 80 ms and 261 ± 57 ms respectively.
This study showed that MRF has the potential to identify high-grade prostate cancers and illustrated the power of a rich quantitative space in characterizing disease. Future applications of prostate MRF includes correlating T1 and T2 values with tumor aggressiveness, guiding targeted biopsies and monitoring patients on active surveillance for prostate cancer. Work on sequence improvement involves generating a 3D volumetric MRF sequence for prostate, obtaining ADC values with MRF and improving the overall spatial resolution so that conventional imaging could potentially be eliminated altogether.
Abdomen
Adopting MRF for abdominal imaging comes with its own unique challenges and necessitates modifying the original MRF sequence. Challenges included developing a fast sequence that could yield the needed data in a single breath hold, provide a high spatial resolution, cover the wide range of tissues within the abdomen and compensate for both B0 and B1 inhomogeneities. A couple of approaches have been proposed for imaging abdomen using the MRF framework[7,16].
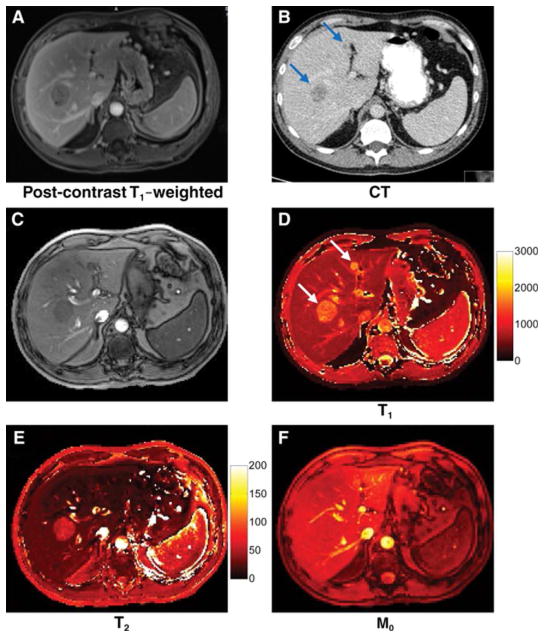
Chen et al[16] adopted an approach where the authors measured B1 variation through a separate scan based on the Bloch-Seigert method[52] and incorporated the corresponding values in the dictionary simulation to reduce the effects of B1 inhomogeneity on tissue properties. Additionally, a SSFP based MRF acquisition was used due to its robustness to B0 inhomogeneity. This sequence was validated for abdomen MRF in asymptomatic subjects and the obtained T1 and T2 values of various abdominal organs were congruent with literature[16]. The clinical utility of this sequence was also tested in 6 patients with 20 metastatic adenocarcinoma lesions to the liver. The mean T1 and T2 in metastases (mean,1673 msec ± 331, 43 msec ± 13,), were significantly different from surrounding liver parenchyma (mean, 840 msec ± 113, 28 msec ± 3; P < .0001 and P < .01) and healthy liver parenchyma in volunteers respectively (mean, 745 msec ± 65, 31 msec ± 6; P < .0001 and P = .021)[53](Figure 4). The total scan time (including the time for B1 mapping) for this acquisition was 19 seconds per slice for a resolution of 1.9 × 1.9 × 5 mm3[16].
Figure 4. MRF in liver metastases.
(A) Post-contrast T1 weighted (A) and contrast enhanced CT image (B) show two liver metastases from lung adenocarcinoma. (C) shows a MRF image summed from all undersampled images. (D–F) show MRF-derived quantitative T1, T2 and M0 maps. The T1, T2 and M0 values of these lesions were 1462 and 57 ms and 1582 and 19 ms respectively, while the surrounding liver parenchyma had T1 and T2 values of 686 and 26 ms respectively.
Reprinted with permission from Radiology Society of North America (RSNA) from Figure 5, Chen Y, Jiang Y, Pahwa S, Ma D, Lu L, Twieg MD, Wright KL, Seiberlich N, Griswold MA, Gulani V: MR Fingerprinting for Rapid Quantitative Abdominal Imaging. Radiology 2016, 279:278–286 [19].
Other groups have also shown approaches to overcome B1 inhomogeneity[6,7,17]. Cloos, et al[7] proposed an alternative approach to B1 inhomogeneity (shown in simulation in the abdomen and demonstrated in vivo for high field musculoskeletal imaging as below) where the combination mode of the RF transmit coils or “B1 illumination mode” was used as an additional MRF acquisition parameter. In this method, similar to other acquisition parameters such as FA, TR, etc. the B1 illumination mode was varied from time point to time point to generate temporal variation in the B1 inhomogeneity of the fingerprinting signal. Using the MRF framework, they thus reduced the effects of B1 inhomogeneity on the estimated tissue properties. A combination of SSFP and spoiled gradient echo based modules were used to reduce the effects of B0 inhomogeneity on the estimated tissue properties.
Musculoskeletal
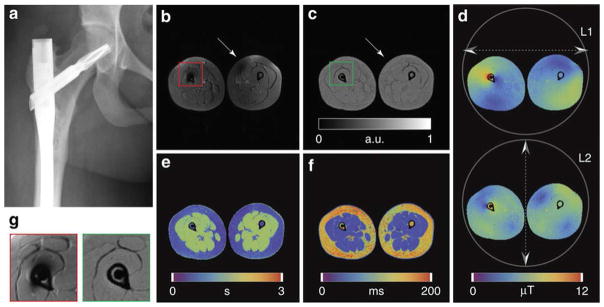
MRI forms a vital cog in musculoskeletal (MSK) imaging, with its ability to provide superior soft-tissue contrast and differentiate between the wide variety of tissues seen in musculoskeletal imaging, namely bone marrow, muscle, fat, cartilage, tendons and soft tissues. Imaging in the presence of orthopedic implants is a challenging aspect of MRI because the images are affected by variations in RF fields and implants lead to areas of signal loss. The MRF approach to overcome B1 inhomogeneity, as proposed by Cloos, et al[7] was successful for obtaining tissue maps devoid of shading artifacts despite the presence of orthopedic implants(Figure 5). These maps were generated with resolution of 1.4 × 1.4 × 5 mm3 and total scan time of 8 minutes 18 seconds for 18 slices.
Figure 5. Conventional MRI versus Plug-and-Play (PnP)-MRF in the presence of an orthopedic implant.
(A) X-ray image showing the orthopedic implant. (B) Contrast weighted axial image through the legs obtained using a conventional inversion recovery (IR) TSE sequence. Signal voids appear both in the vicinity of the implant and in the contralateral leg. (C–F) Quantitative maps obtained using PnP-MRF, including proton density (PD), B10, T1 and T2, respectively. Units are arbitrary for PD (C), micro Tesla for B10 (D), seconds for T1 (E) and milliseconds for T2 (F). (G) Enlargements, extracted from B (red frame) and C (green frame), comparing the region surrounding the implant with conventional (left) and PnP-MRF (right) approaches. Note the absence of B1 -related signal voids in any of the PnP-MRF parameter maps.
Reprinted under Creative Commons License from Figure 6, Cloos MA, Knoll F, Zhao T, Block KT, Bruno M, Wiggins GC, Sodickson DK: Multiparametric imaging with heterogeneous radiofrequency fields. Nat. Commun. 2016, 7:12445. doi:10.1038/ncomms12445[7].
Cardiac
Myocardial tissue property mapping has come into focus in cardiac imaging for the ability to detect pathology earlier than conventional cardiac imaging. Cardiac MRF as described by Hamilton, et al[15] provided simultaneous estimation of T1, T2 and M0 values with a resolution of 1.6 × 1.6 × 8 mm3 using a breath-hold acquisition of 16 heartbeats. While there are no patient data available yet, comparison of T1 and T2 MRF values from normal volunteers show good concordance with conventional mapping methods[15]. Further technical development involves reducing the scan time, volumetric acquisition for whole heart coverage and optimizing the M0 values obtained.
Microvascular structure
The MRF framework has also been extended to characterize the microvascular properties termed MR vascular fingerprinting (MRvF). Christen, et al[10] described a new MRF approach to measure microvascular properties such as cerebral blood volume (CBV), mean vessel radius (R) and the blood oxygenation saturation (SO2). The acquisition sequence was based on a gradient echo sampling of the free induction decay and spin echo (GEFSIDE) sequence. The acquisition was performed before and 2 minutes after injection of an iron-based contrast agent (ferumoxytol). The dictionary was simulated using a different mathematical model both before and after contrast injection and the ratio of the pre and post contrast signal evolutions was used as the fingerprint[54]. The best fit between the observed fingerprint and the dictionary was determined by using least squares method to generate CBV, R and SO2 for voxels. Pre and post contrast scans were acquired using a resolution of 1.6 × 1.6 × 1.5 mm3 with scan time of 4 minutes for whole brain coverage[10]. Preliminary data showed high contrast in CBV maps between grey and white matter, suggesting differential perfusion. R and SO2 maps were more homogenous[10](Figure 6). Other authors have further built upon this foundation and applied MRvF in rats studies to either get more realistic vascular maps[12] or evaluate differences in microvascular properties in strokes and brain tumors[11]. Lemasson, et al[11]tested the MRvF technique in 115 rats divided into three models, namely brain tumors, strokes and healthy animals, to obtain maps of the microvascular architecture (i.e., blood volume fraction, vessel diameter) and function (blood oxygenation) simultaneously. These findings were compared with conventional MR approaches and histopathology analysis for validation. MRvF could robustly distinguish between healthy and pathological brain tissues with different behaviors in tumor and stroke models. MRvF also showed that two pathologically different brain tumors had distinct microvascular signal evolutions. This in-vivo estimation of microvascular properties, if efficiently translated to humans, can potentially improve diagnosis and management of brain diseases.
Figure 6. Representative MR Vascular fingerprinting (MRvF) maps.
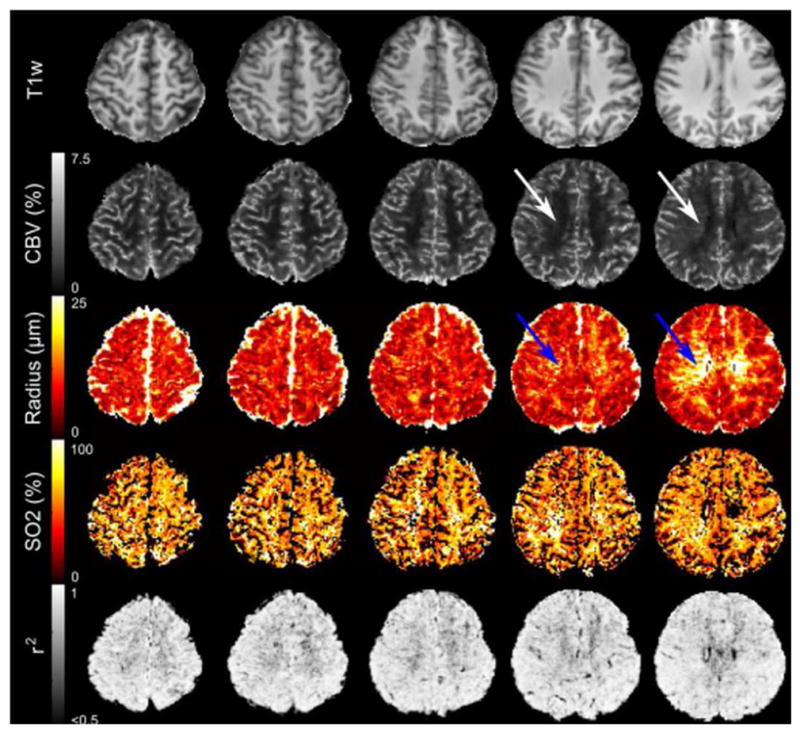
Representative parametric maps for T1, cerebral blood volume (CBV), Radius and oxygen saturation (SO2) obtained from normal volunteer who had the highest correlation coefficient (r2) for the match between the fingerprint and the dictionary. White and blue arrows indicate that the technique is sensitive enough to detect the relatively small medullary veins of the deep white matter.
Reprinted with permission from Elsevier from Figure 5, Christen T, Pannetier N, Ni W, Qiu D, Moseley M, Schuff N, Zaharchuk G: MR Vascular Fingerprinting: A New Approach to Compute Cerebral Blood Volume, Mean Vessel Radius, and Oxygenation Maps in the Human Brain. NeuroImage 2014, 89:262–270[10].
Perfusion (MRF-ASL)
ASL is a promising non-contrast MR perfusion technique. Since, the ASL signal is affected by multiple hemodynamic parameters, both Wright et al[KL Wright et al., Abstract 0417.22nd Annual Meeting of International Society of Magnetic Resonance in Medicine, Milan, Italy, 2014] and Su, et al[9] recently explored the feasibility of developing an ASL technique based on MRF framework to simultaneously get multiple hemodynamic parameters with one scan. The MRF-ASL acquisition was performed using an EPI readout with whole brain coverage in 3 minutes of scan time and a resolution of 2.8 × 2.8 × 10 mm3[9]. Different hemodynamic parameters were extracted depending on the mathematical model used and compared with dictionary values. The sequence was validated in volunteers using a hypercapnia challenge, which showed expected increase in perfusion in response to CO2 stimulation, and in patients with Moya-Moya disease, which showed delay in bolus arrival time on the side with the stenotic internal carotid artery.
Conclusion
MRF heralds a new approach to quantitative MRI by providing a framework to measure several tissue properties in a single and time-efficient acquisition. Various sequences can be tailored to this MRF framework depending on the specific tissue properties that need to be measured, including T1, T2, M0, perfusion parameters, B0, B1, etc. Current MRF sequences have been shown to be repeatable[2,13,14]. MRF offers the possibility of bringing quantitative imaging into clinical routine, potentially allowing more precise and automated diagnosis and easier comparison between data acquired in different scans, at different times and in different locations. Follow-up studies and patient specific therapy management can also benefit from such a quantitative approach. In the future, MRF could therefore play a role in the development of imaging biomarkers and novel therapies. Improvements in sequence design, efficiency, acquisition speed, post-processing steps and image quality are continuous and on-going processes. Clinical validation of MRF requires the establishment of inter-disciplinary biomedical engineering and clinical teams that can simultaneously develop new hypotheses and test the technology in various clinical applications and organ systems. This can help MRF become an effective clinical tool and facilitate development of a rapid, “one-scan, multiple-property” approach to quantitative MR imaging.
Highlights.
MRF is a novel approach to quantitative MRI that allows measurement of multiple tissue properties in a single, time-efficient acquisition.
MRF acquisition relies on deliberately varying MR system parameters such that each tissue produces a unique single evolution called “fingerprint”.
Pattern matching the tissue fingerprint to a database of fingerprints helps to visualize specific tissue properties.
Some of the tissue properties measured include longitudinal relaxation time (T1), transverse relaxation time (T2), proton density (M0) and off-resonance frequency (B0).
Clinically MRF has been used to differentiate between various brain tumors, separate prostate cancer from normal prostatic tissue and characterize liver metastases.
Development and validation of MRF technology and testing new clinical applications remain continuous and ongoing processes.
Acknowledgments
Funding source: The authors receive research support from Siemens Healthcarand NIH. The research work presented is funded by National Institutes of Health grants 1R01EB016728, 1R01BB017219, 1R01DK098503, 1R01CA208236, 2R01HL094557, 1R21EB021562 and 1R01EB017219-01)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kransdorf MJ, Murphey MD. Radiologic evaluation of soft-tissue masses: a current perspective. AJR Am J Roentgenol. 2000;175:575–587. doi: 10.2214/ajr.175.3.1750575. [DOI] [PubMed] [Google Scholar]
- 2**.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature. 2013;495:187–192. doi: 10.1038/nature11971. The paper that first introduced the framework and showed proof-of-principle implementation of MR Fingerprinting (MRF). The authors used an inversion recovery prepared balanced steady state free precession (IR-bSSFP) for the MRF framework and showed MRF to be accurate, precise and intolerant to undersampling and motion errors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connor JPB, Aboagye EO, Adams JE, Aerts HJWL, Barrington SF, Beer AJ, Boellaard R, Bohndiek SE, Brady M, Brown G, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14:169–186. doi: 10.1038/nrclinonc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coppo S, Mehta BB, McGivney D, Ma D, Chen Y, Jiang Y, et al. Overview of magnetic resonance fingerprinting. [date unknown] [Google Scholar]
- 5.Radiology (ESR) ES of: Magnetic Resonance Fingerprinting -a promising new approach to obtain standardized imaging biomarkers from MRI. Insights Imaging. 2015;6:163–165. doi: 10.1007/s13244-015-0403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buonincontri G, Sawiak SJ. MR fingerprinting with simultaneous B1 estimation. Magn Reson Med. 2016;76:1127–1135. doi: 10.1002/mrm.26009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Cloos MA, Knoll F, Zhao T, Block KT, Bruno M, Wiggins GC, Sodickson DK. Multiparametric imaging with heterogeneous radiofrequency fields. Nat Commun. 2016:7. doi: 10.1038/ncomms12445. The authors described an approach to overcome heterogeneous radiofrequency transmit fields (B1). This was termed as Plug and Play (PnP) MRF and was tested under high-field strengths and with orthopedic implants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rieger B, Zimmer F, Zapp J, Weingärtner S, Schad LR. Magnetic resonance fingerprinting using echo-planar imaging: Joint quantification of T1 and T2* relaxation times. Magn Reson Med. 2016 doi: 10.1002/mrm.26561. [DOI] [PubMed] [Google Scholar]
- 9.Su P, Mao D, Liu P, Li Y, Pinho MC, Welch BG, Lu H. Multiparametric estimation of brain hemodynamics with MR fingerprinting ASL. Magn Reson Med. 2016 doi: 10.1002/mrm.26587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Christen T, Pannetier N, Ni W, Qiu D, Moseley M, Schuff N, Zaharchuk G. MR Vascular Fingerprinting: A New Approach to Compute Cerebral Blood Volume, Mean Vessel Radius, and Oxygenation Maps in the Human Brain. NeuroImage. 2014;89:262–270. doi: 10.1016/j.neuroimage.2013.11.052. The authors showed how the MRF framework could be extended to mapping microvascular properties using a new and different mathematical model to generate the dictionary. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemasson B, Pannetier N, Coquery N, Boisserand LSB, Collomb N, Schuff N, Moseley M, Zaharchuk G, Barbier EL, Christen T. MR Vascular Fingerprinting in Stroke and Brain Tumors Models. Sci Rep. 2016;6:37071. doi: 10.1038/srep37071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pouliot P, Gagnon L, Lam T, Avti PK, Bowen C, Desjardins M, Kakkar AK, Thorin E, Sakadzic S, Boas DA, et al. Magnetic resonance fingerprinting based on realistic vasculature in mice. NeuroImage. 2017;149:436–445. doi: 10.1016/j.neuroimage.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Jiang Y, Ma D, Keenan KE, Stupic KF, Gulani V, Griswold MA. Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom. Magn Reson Med. 2017;78:1452–1457. doi: 10.1002/mrm.26509. The authors scanned phantoms over 34 consecutive days and showed that MRF measurements of T1 and T2 were highly reproducible over time and across wide ranges of T1 and T2 values, thereby showing that MRF is a robust technique. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR Fingerprinting Using Fast Imaging with Steady State Precession (FISP) with Spiral Readout. Magn Reson Med. 2015;74:1621–1631. doi: 10.1002/mrm.25559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Hamilton JI, Jiang Y, Chen Y, Ma D, Lo W-C, Griswold M, Seiberlich N. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn Reson Med. 2017;77:1446–1458. doi: 10.1002/mrm.26216. The authors showed extended MRF framework for cardiac imaging. Cardiac imaging is a challenge due to its dynamic nature and necessity for fast imaging but the authors were able to generate single-slice T1, T2, and M0 maps of the heart within a single breath-hold. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Chen Y, Jiang Y, Pahwa S, Ma D, Lu L, Twieg MD, Wright KL, Seiberlich N, Griswold MA, Gulani V. MR Fingerprinting for Rapid Quantitative Abdominal Imaging. Radiology. 2016;279:278–286. doi: 10.1148/radiol.2016152037. The authors described another method to overcome B1 inhomogeneity by acquiring a separate B1 scan before implementing SSFP based MRF framework. The utility of this method in abdominal imaging was validated in both normal human volunteers and patients with focal liver lesions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonincontri G, Schulte RF, Cosottini M, Tosetti M. Spiral MR fingerprinting at 7 T with simultaneous B1 estimation. Magn Reson Imaging. 2017;41:1–6. doi: 10.1016/j.mri.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Assländer J, Glaser SJ, Hennig J. Pseudo Steady-State Free Precession for MR-Fingerprinting. Magn Reson Med. 2017;77:1151–1161. doi: 10.1002/mrm.26202. [DOI] [PubMed] [Google Scholar]
- 19.Anderson CE, Donnola SB, Jiang Y, Batesole J, Darrah R, Drumm ML, Brady-Kalnay SM, Steinmetz NF, Yu X, Griswold MA, et al. Dual Contrast - Magnetic Resonance Fingerprinting (DC-MRF): A Platform for Simultaneous Quantification of Multiple MRI Contrast Agents. Sci Rep. 2017;7:8431. doi: 10.1038/s41598-017-08762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson CE, Wang CY, Gu Y, Darrah R, Griswold MA, Yu X, Flask CA. Regularly incremented phase encoding - MR fingerprinting (RIPE-MRF) for enhanced motion artifact suppression in preclinical cartesian MR fingerprinting. Magn Reson Med. 2017 doi: 10.1002/mrm.26865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, Chen Y, Ma D, Jiang Y, Herrmann KA, Vincent JA, Dell KM, Drumm ML, Brady-Kalnay SM, Griswold MA, et al. Preclinical MR fingerprinting (MRF) at 7 T: effective quantitative imaging for rodent disease models. NMR Biomed. 2015;28:384–394. doi: 10.1002/nbm.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt P, Griswold MA, Jakob PM, Kotas M, Gulani V, Flentje M, Haase A. Inversion recovery TrueFISP: quantification of T(1), T(2), and spin density. Magn Reson Med. 2004;51:661–667. doi: 10.1002/mrm.20058. [DOI] [PubMed] [Google Scholar]
- 23.Ehses P, Seiberlich N, Ma D, Breuer FA, Jakob PM, Griswold MA, Gulani V. IR TrueFISP with a golden-ratio-based radial readout: fast quantification of T1, T2, and proton density. Magn Reson Med. 2013;69:71–81. doi: 10.1002/mrm.24225. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt P, Griswold MA, Gulani V, Haase A, Flentje M, Jakob PM. A simple geometrical description of the TrueFISP ideal transient and steady-state signal. Magn Reson Med. 2006;55:177–186. doi: 10.1002/mrm.20738. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Ma D, Jerecic R, Duerk J, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using the quick echo splitting NMR imaging technique. Magn Reson Med. 2017;77:979–988. doi: 10.1002/mrm.26173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Ma D, Pierre EY, Jiang Y, Schluchter MD, Setsompop K, Gulani V, Griswold MA. Music-based magnetic resonance fingerprinting to improve patient comfort during MRI examinations. Magn Reson Med. 2016;75:2303–2314. doi: 10.1002/mrm.25818. In this paper, the authors showed how acoustic noise during MR scanning could be decreased by using waveforms of music audio files to vary timings of acquisition parameters, such that musical sounds could be produced while simultaneously quantifying multiple tissue properties. This improved patient comfort and at the same time, the tissue values obtained were consistent with those in literature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierre EY, Ma D, Chen Y, Badve C, Griswold MA. Multiscale reconstruction for MR fingerprinting. Magn Reson Med. 2016;75:2481–2492. doi: 10.1002/mrm.25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye H, Ma D, Jiang Y, Cauley SF, Du Y, Wald LL, Griswold MA, Setsompop K. Accelerating magnetic resonance fingerprinting (MRF) using t-blipped simultaneous multislice (SMS) acquisition. Magn Reson Med. 2016;75:2078–2085. doi: 10.1002/mrm.25799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye H, Cauley SF, Gagoski B, Bilgic B, Ma D, Jiang Y, Du YP, Griswold MA, Wald LL, Setsompop K. Simultaneous multislice magnetic resonance fingerprinting (SMS-MRF) with direct-spiral slice-GRAPPA (ds-SG) reconstruction. Magn Reson Med. 2017;77:1966–1974. doi: 10.1002/mrm.26271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao B, Setsompop K, Ye H, Cauley SF, Wald LL. Maximum Likelihood Reconstruction for Magnetic Resonance Fingerprinting. IEEE Trans Med Imaging. 2016;35:1812–1823. doi: 10.1109/TMI.2016.2531640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies M, Puy G, Vandergheynst P, Wiaux Y. A Compressed Sensing Framework for Magnetic Resonance Fingerprinting. SIAM J Imaging Sci. 2014;7:2623–2656. [Google Scholar]
- 32.Jiang Y, Ma D, Bhat H, Ye H, Cauley SF, Wald LL, Setsompop K, Griswold MA. Use of pattern recognition for unaliasing simultaneously acquired slices in simultaneous multislice MR fingerprinting. Magn Reson Med. 2016 doi: 10.1002/mrm.26572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Assländer J, Cloos MA, Knoll F, Sodickson DK, Hennig J, Lattanzi R. Low rank alternating direction method of multipliers reconstruction for MR fingerprinting. Magn Reson Med. 2017 doi: 10.1002/mrm.26639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao X, Liao C, Wang Z, Chen Y, Ye H, He H, Zhong J. Robust sliding-window reconstruction for Accelerating the acquisition of MR fingerprinting. Magn Reson Med. 2017;78:1579–1588. doi: 10.1002/mrm.26521. [DOI] [PubMed] [Google Scholar]
- 35.Cline CC, Chen X, Mailhe B, Wang Q, Pfeuffer J, Nittka M, Griswold MA, Speier P, Nadar MS. AIR-MRF: Accelerated iterative reconstruction for magnetic resonance fingerprinting. Magn Reson Imaging. 2017;41:29–40. doi: 10.1016/j.mri.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Doneva M, Amthor T, Koken P, Sommer K, Börnert P. Matrix completion-based reconstruction for undersampled magnetic resonance fingerprinting data. Magn Reson Imaging. 2017;41:41–52. doi: 10.1016/j.mri.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Zhao B, Setsompop K, Adalsteinsson E, Gagoski B, Ye H, Ma D, Jiang Y, Ellen Grant P, Griswold MA, Wald LL. Improved magnetic resonance fingerprinting reconstruction with low-rank and subspace modeling. Magn Reson Med. 2017 doi: 10.1002/mrm.26701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Li H, Zhang Q, Yuan J, Wang X. Magnetic Resonance Fingerprinting with compressed sensing and distance metric learning. Neurocomputing. 2016;174:560–570. [Google Scholar]
- 39.Zhang X, Zhou Z, Chen S, Chen S, Li R, Hu X. MR fingerprinting reconstruction with Kalman filter. Magn Reson Imaging. 2017;41:53–62. doi: 10.1016/j.mri.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Bloch F. The Principle of Nuclear Induction. Science. 1953;118:425–430. doi: 10.1126/science.118.3068.425. [DOI] [PubMed] [Google Scholar]
- 41.Weigel M, Schwenk S, Kiselev VG, Scheffler K, Hennig J. Extended phase graphs with anisotropic diffusion. J Magn Reson San Diego Calif 1997. 2010;205:276–285. doi: 10.1016/j.jmr.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 42.McGivney DF, Pierre E, Ma D, Jiang Y, Saybasili H, Gulani V, Griswold MA. SVD compression for magnetic resonance fingerprinting in the time domain. IEEE Trans Med Imaging. 2014;33:2311–2322. doi: 10.1109/TMI.2014.2337321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cauley SF, Setsompop K, Ma D, Jiang Y, Ye H, Adalsteinsson E, Griswold MA, Wald LL. Fast group matching for MR fingerprinting reconstruction. Magn Reson Med. 2015;74:523–528. doi: 10.1002/mrm.25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bojorquez JZ, Bricq S, Acquitter C, Brunotte F, Walker PM, Lalande A. What are normal relaxation times of tissues at 3 T? Magn Reson Imaging. 2017;35:69–80. doi: 10.1016/j.mri.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 45.de Bazelaire CMJ, Duhamel GD, Rofsky NM, Alsop DC. MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3. 0 T: preliminary results. Radiology. 2004;230:652–659. doi: 10.1148/radiol.2303021331. [DOI] [PubMed] [Google Scholar]
- 46.Wansapura JP, Holland SK, Dunn RS, Ball WS. NMR relaxation times in the human brain at 3. 0 tesla. J Magn Reson Imaging JMRI. 1999;9:531–538. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 47.Badve C, Yu A, Rogers M, Ma D, Liu Y, Schluchter M, Sunshine J, Griswold M, Gulani V. Simultaneous T1 and T2 Brain Relaxometry in Asymptomatic Volunteers using Magnetic Resonance Fingerprinting. Tomogr J Imaging Res. 2015;1:136–144. doi: 10.18383/j.tom.2015.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badve C, Yu A, Dastmalchian S, Rogers M, Ma D, Jiang Y, Margevicius S, Pahwa S, Lu Z, Schluchter M, et al. MR Fingerprinting of Adult Brain Tumors: Initial Experience. AJNR Am J Neuroradiol. 2017;38:492–499. doi: 10.3174/ajnr.A5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma D, Jiang Y, Chen Y, McGivney D, Mehta B, Gulani V, Griswold M. Fast 3D magnetic resonance fingerprinting for a whole-brain coverage. Magn Reson Med. doi: 10.1002/mrm.26886. [date unknown] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao C, Bilgic B, Manhard MK, Zhao B, Cao X, Zhong J, Wald LL, Setsompop K. 3D MR fingerprinting with accelerated stack-of-spirals and hybrid sliding-window and GRAPPA reconstruction. NeuroImage. 2017;162:13–22. doi: 10.1016/j.neuroimage.2017.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu AC, Badve C, Ponsky LE, Pahwa S, Dastmalchian S, Rogers M, Jiang Y, Margevicius S, Schluchter M, Tabayoyong W, et al. Development of a Combined MR Fingerprinting and Diffusion Examination for Prostate Cancer. Radiology. 2017;283:729–738. doi: 10.1148/radiol.2017161599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sacolick LI, Wiesinger F, Hancu I, Vogel MW. B1 mapping by Bloch-Siegert shift. Magn Reson Med. 2010;63:1315–1322. doi: 10.1002/mrm.22357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Jiang Y, Pahwa S, Ma D, Lu L, Twieg MD, Wright KL, Seiberlich N, Griswold MA, Gulani V. MR Fingerprinting for Rapid Quantitative Abdominal Imaging. Radiology. 2016;279:278–286. doi: 10.1148/radiol.2016152037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pannetier NA, Sohlin M, Christen T, Schad L, Schuff N. Numerical modeling of susceptibility-related MR signal dephasing with vessel size measurement: phantom validation at 3T. Magn Reson Med. 2014;72:646–658. doi: 10.1002/mrm.24968. [DOI] [PMC free article] [PubMed] [Google Scholar]