Figure 7. High-throughput proteomic analysis confirms CAG length-dependent changes in 6-month striatum of HD mice.
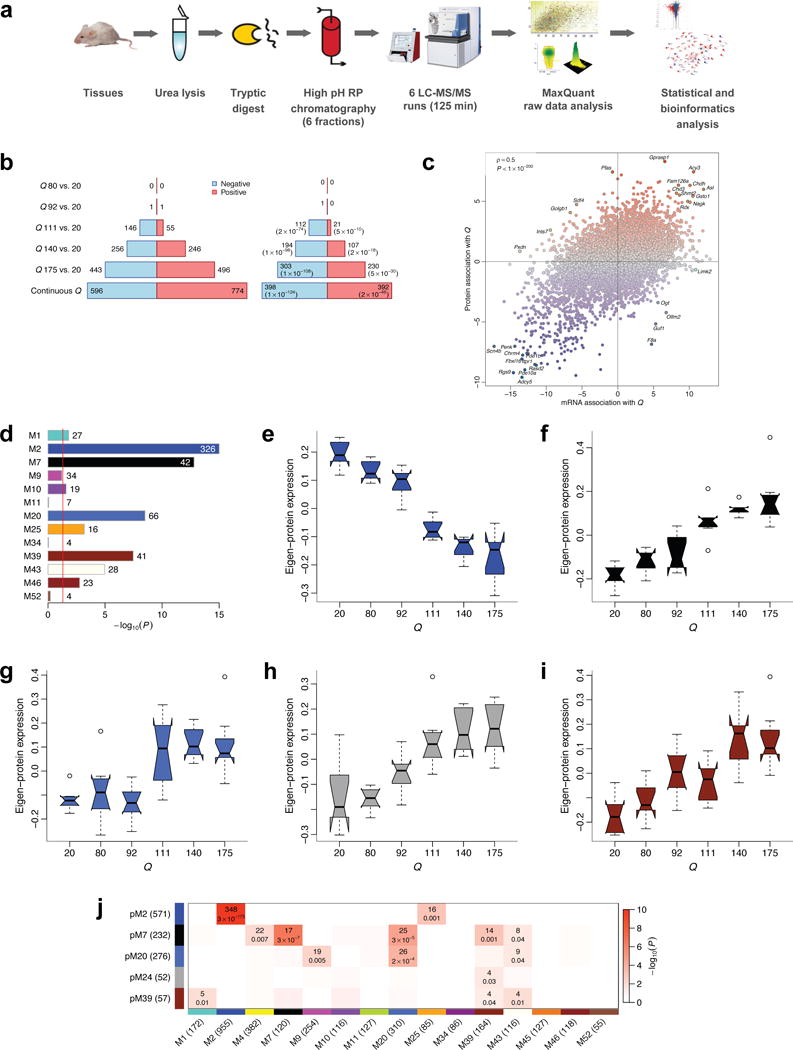
(a) Workflow of proteomic profiling. (b) Left barplot represents numbers of proteins significantly (FDR<0.1) associated with CAG length (Q). Blue (red) bars denote proteins down- (up-) regulated with increasing CAG length. Right barplot represents numbers of significant proteins whose mRNA also changes significantly (FDR<0.1) and in the same direction. Values in brackets are hypergeometric p-values of overlaps of significant protein and gene mRNA profiles. (c) Z statistics for protein association with Q vs. the corresponding mRNA Z statistics. Blue (red) dots represent proteins whose abundance decreases (increases) with increasing CAG length. Selected concordant and discordant genes are labeled. (d) Bars show hypergeometric enrichment p-values of mRNA module genes in proteins that are significantly differentially abundant in the same direction as the module. Numbers give the corresponding gene counts. For clarity, the p-value axis is truncated to 10−15. (e-i) Summary profiles of the 5 protein network modules with strongest association with Q, as a function of Q. Boxes indicate the median, interquartile range and confidence interval for the median. Whiskers indicate the range of data up to 1.5 of the inter-quartile range; points beyond the range of whiskers (if any) are shown individually. (j). Numbers of common genes and hypergeometric overlap p-values among selected protein (rows) and mRNA (columns) modules. Row and column labels indicate module sizes within the 7,039 genes common to both mRNA and protein network analyses.
