Extended Data Fig. 4. RT027 strains have a L172I mutation at a highly conserved site.
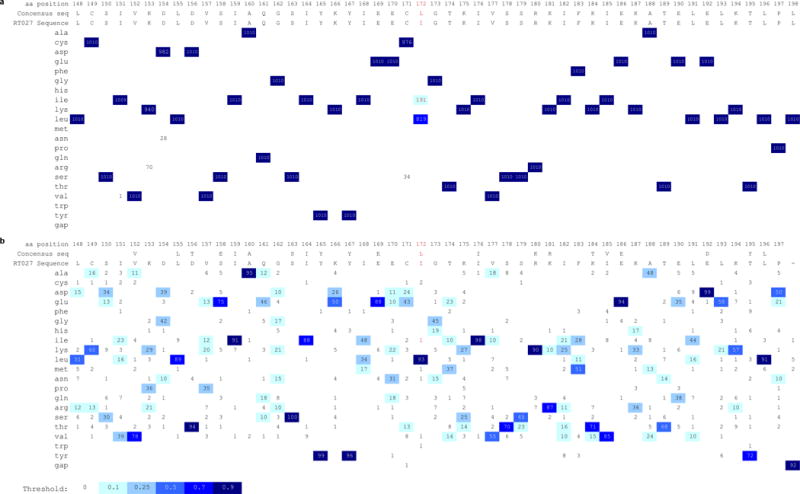
a, treR genes from available C. difficile WGS files on NCBI (accessed 11-May-2017) were identified by tblastn and translated to protein sequences. Sequence fragments < 240 amino acids were discarded and the remaining 1010 sequences aligned with Clustal Omega37. All 191 sequences containing the L172I SNP also contained the thyA gene, a marker for the RT027 lineage. ThyA was not found in any other genomes. Numbers indicate number of sequences with corresponding amino acid in that position. Multiple sequence alignment visualization generated with ProfileGrid.38 b, The TreR protein sequence from RT027 strain R20291 was blasted against non-C. difficile sequences in the NCBI database and the top 99 matches (along with R20291treR) aligned with Clustal Omega. The Leucine at position 172 was found to be conserved in 93 of 99 non-C. difficile sequences. To confirm the importance of this residue, TreR was blasted against all non-Clostridial sequences in the NCBI database and the top 500 hits saved. Following removal of duplicate species 191 sequences were aligned with Clustal Omega. The Leucine at residue 172 was conserved in 83% of sequences (not shown).
