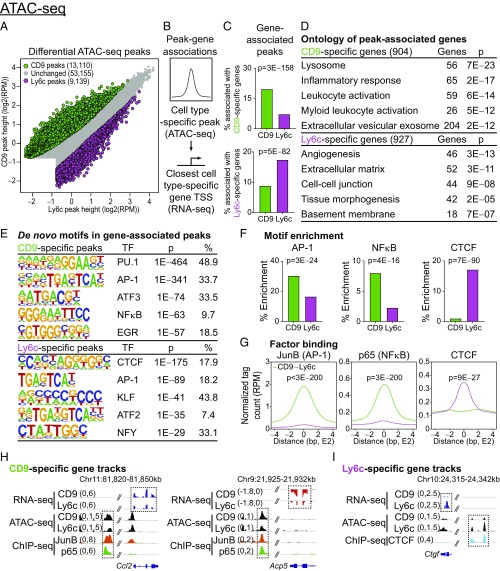
Fig. 4.
CD9 ATMs have an inflammatory chromatin landscape driven by activating transcription factors. (A) Differential analysis of ATAC-seq peaks from CD9 or Ly6c ATMs of HFD-fed B6 mice [n = 3 per arm; fold change > 1.5, false discovery rate < 0.05, and >0.5 reads assigned per million mapped reads (RPM)]. (B) Diagram describing association between cell type-specific peaks and closest cell type-specific gene [within 50 kb of the transcription start site (TSS)]. (C) Percent of ATAC-seq peaks associated with CD9- or Ly6c-specific genes (P values determined by Fisher’s exact test). (D) Gene ontology for cell type-specific genes, associated with corresponding cell-type specific peaks. Most significant, nonredundant biologic process, molecular function, or cellular component terms with gene number and adjusted P value are shown. (E) De novo motif search within ATAC-seq peaks associated with a corresponding cell type-specific gene [consensus motif, transcription factor (TF), P value, and percentage of targets shown]. (F) Percent enrichment of AP-1, NF-κB, and CTCF motifs within CD9- or Ly6c-specific peaks (P values determined y Fisher’s exact test). (G) Occupancy of JunB (AP-1 subunit) and CTCF at CD9- or Ly6c-specific regions of open chromatin (P values determined by Wilcoxon test). (H and I) Representative RNA-seq, ATAC-seq, and ChIP-seq browser tracks displaying CD9- (H) or Ly6c-specific (I) gene loci. Legacy ChIP-seq tracks for JunB (AP-1) or p65 (NF-κB) in lipopolysaccharide-treated BMDM or CTCF in untreated BMDM are displayed. Representative of two or more independent experiments.