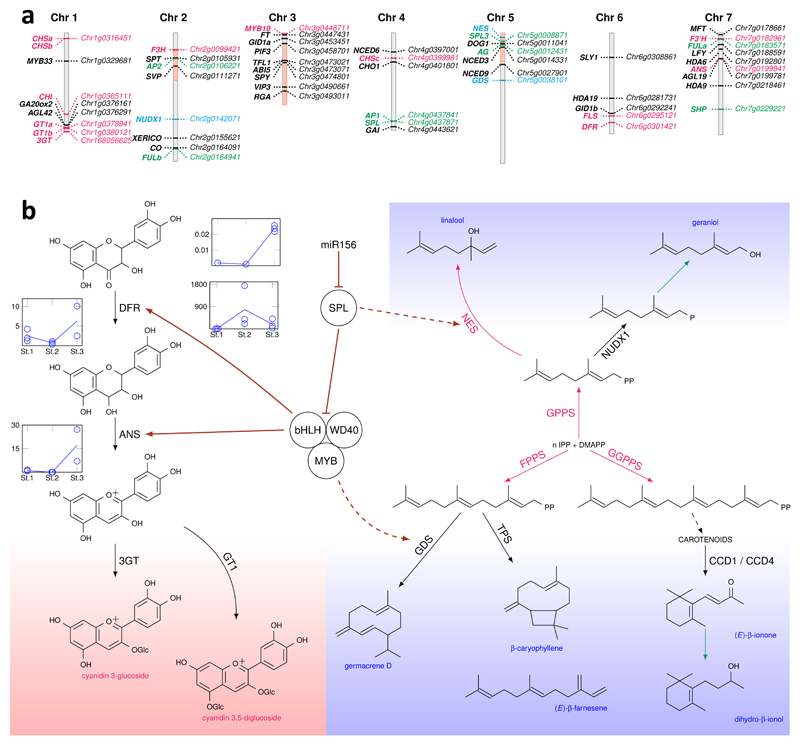
Figure 3. Inter-regulatory connections between color biosynthesis and some scent pathways.
a, Schematic representation of the rose chromosomes together with the position of candidate genes for anthocyanin pigments and volatile molecules biosynthesis and for flowering. Chromosome segments 2.4, 3.2-3.6 and 5.1 originating only from R. chinensis are indicated in light red. Anthocyanin synthesis genes are indicated in red; terpene biosynthesis genes in blue; flowering time genes in black; and development genes in green.
b, Schematic representation of interconnections between color (pink background) and scent (blue background) pathways. Gene expression data show the anti-correlation between miR156 and SPL9 genes during petal development. RT-qPCR was performed on petals harvested at three successive stages: Non-colored petals early during development (St1); Petals at onset of anthocyanin synthesis (St2); Fully colored petals (St3).
Black arrows: biosynthetic steps reported in the rose. Red arrows: biosynthetic steps reported in other species, but not in the rose. Green arrows: putative steps with unknown enzymes. Dashed black arrow: Several enzymatic steps. Maroon arrows: Gene regulation reported in A. thaliana, but not in the rose. Dashed maroon arrow: putative gene regulations. IPP: isopentenyl diphosphate, DMAPP: dimethylallyl diphosphate, DFR: dihydroflavonol-4-reductase, ANS: anthocyanidin synthase, 3GT: anthocyanidin 3-O-glucosyltransferase, GT1: anthocyanidin 3,5-diglucosyltransferase, GPPS: geranyl diphosphate synthase, FPPS: farnesyl diphosphate synthase, GGPPS: geranylgeranyl diphosphate synthase, GDS: germacrene D synthase, TPS: terpene synthase, NES: linalool/nerolidol synthase, CCD1/4 : carotenoid cleavage dioxygenases 1/4, NUDX1: nudix hydrolase1.