Abstract
Many neuroscience questions center around understanding how the molecules and wiring in neural circuits mechanistically yield behavioral functions, or go awry in disease states. However, mapping the molecules and wiring of neurons across the large scales of neural circuits has posed a great challenge. We recently developed expansion microscopy (ExM), a process in which we physically magnify biological specimens such as brain circuits. We synthesize throughout preserved brain specimens a dense, even mesh of a swellable polymer such as sodium polyacrylate, anchoring key biomolecules such as proteins and nucleic acids to the polymer. After mechanical homogenization of the specimen-polymer composite, we add water, and the polymer swells, pulling biomolecules apart. Due to the larger separation between molecules, ordinary microscopes can then perform nanoscale resolution imaging. We here review the ExM technology as well as applications to the mapping of synapses, cells, and circuits, including deployment in species such as Drosophila, mouse, non-human primate, and human.
Introduction
At the core of many neuroscience questions, ranging from understanding how memories are encoded, to how neurons transform sensory inputs into motor outputs, to how emotions and decisions are implemented, is a need to understand how neural circuits are organized to yield complex emergent functions. Understanding the nature of brain disorders, and pointing the way to new therapeutics, is also increasingly demanding a knowledge of how brain cells, molecular cascades, and connections change in disease states. Ideally one would be able to map biomolecules such as neurotransmitters, receptors, and ion channels, across the spatial extents of neurons and neural circuits. Traditional microscopes are limited by diffraction, and thus specialized technologies have been required to perform imaging with nanoscale precision. Electron microscopy is capable of nanoscale resolution, and has yielded many insights into the wiring diagrams of neural circuits12, but typically yields little molecular information about the molecules in those circuits. Super-resolution light microscopy methods have powerfully revealed many molecular features of neurons at the nanoscale level34, but such methods are difficult to apply to extended 3-D specimens, such as neural circuits, due to their speed and complexity. To address the need for a method of imaging extended 3-D objects such as neural circuits, with molecular information, at nanoscale resolution, we recently developed a novel modality of imaging. In contrast to earlier methods of nanoscale imaging that magnify information emitted from a specimen, we physically magnify the specimen itself5.
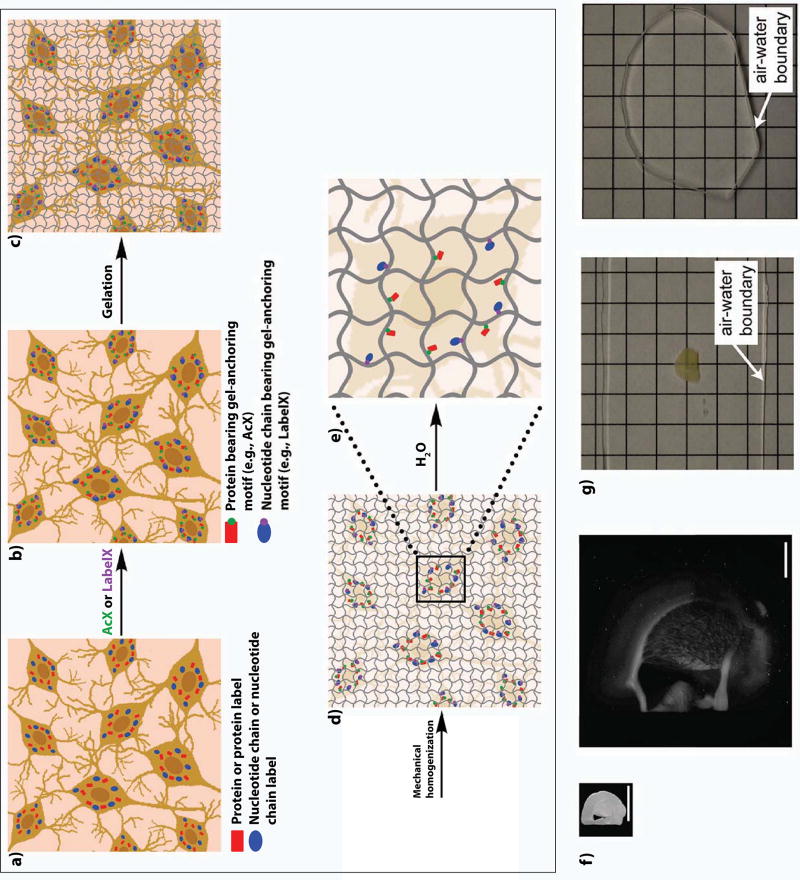
In this new methodology, which we call expansion microscopy (ExM, schematized in Fig. 1a–e), we synthesize a dense, interconnected web of a swellable polymer, such as sodium polyacrylate, throughout a preserved specimen such as a brain specimen. The polymer is very dense, such that the distance between adjacent polymer threads is on the order of the dimension of a biomolecule. We anchor biomolecules such as proteins or RNA, or labels bound to those biomolecules (such as antibodies), to the polymer network via covalently binding anchoring molecules. We treat the specimen with heat, detergent, and/or enzymes to mechanically homogenize the specimen so that it can expand evenly, and then finally we add water. The swellable polymer absorbs the water, and expands, bringing the anchored biomolecules or labels along (Fig. 1f). The net result is that biomolecules or labels that are initially localized within the diffraction limit of a traditional microscope, are now separated in space to distances far enough that they can now be resolved. The specimen also becomes completely transparent, having become mostly water (Fig. 1g).
Figure 1. Expansion microscopy workflow.
Biomolecules, or labels highlighting biomolecules of interest, in fixed cells or tissues (a), are functionalized with chemical handles (AcX, green, binds proteins; LabelX, purple, binds nucleic acids such as mRNA) that enable them to be covalently anchored (b) to a swellable polymer mesh (composed of crosslinked sodium polyacrylate) that is evenly and densely synthesized throughout the specimen (c). The sample is mechanically homogenized by treatment with heat, detergent, and/or proteases (d). Adding water initiates polymer swelling (e), which results in biomolecules or labels being pulled apart from each other in an even, isotropic fashion, and thus enabling nanoscale resolution imaging on conventional microscopes (f, adapted from ref. 5). Expansion significantly reduces scattering of the sample, since the sample is mostly water (g, adapted from ref. 5). In g, a 200 µm thick fixed mouse brain slice is opaque before ExM, but after expansion is completely transparent.
ExM builds from two sets of ideas that go back into the late 1970s and early 1980s. Around that time, the physicist Toyoichi Tanaka at MIT was creating and studying the physics of swellable gels6, and found that they could swell many orders of magnitude in volume in ways that could be precisely described via phase transition mathematics. Around the same time, Peter Hausen and Christine Dreyer at the Max Planck Institute developed polymer hydrogel embedding of fixed tissues for the enhancement of imaging, synthesizing polyacrylamide networks throughout preserved specimens7. ExM fuses these two old concepts to enable physical magnification of specimens, with precision down to the nanoscale.
In this review, we first discuss the principles of how ExM works, discussing some of the rapidly-exploding family of protocols that have been invented in the past few years that are making ExM easier to use and more powerful, and then we discuss some current applications in the field of neuroscience.
Principles of how expansion microscopy works
Since our discovery of expansion microscopy, accompanied by a proof-of-concept protocol and validation data showing its high performance in cultured mammalian cells and mouse brain tissue, published in 20155, we have developed several variants specialized for simple visualization of proteins8 and RNA9 using off-the-shelf chemicals, variants that can expand cells and tissues to much greater extents than the original protocol10, and variants that can easily be applied to human pathology specimens11. Several groups, including ours, have also shown the technology to work in a wide diversity of non-brain tissues, both normal and diseased (e.g., cancer-containing)8,11,12, and even with pathogens like bacteria13. Many other groups have joined in creating expansion microscopy protocol variants as well12,14,15,16. Demonstrations of utility as well as scientific applications to a diversity of neuroscience questions have begun, in a diversity of species ranging from planaria17, to Drosophila18, to mouse19,20, to non-human primate8, to human21. Rather than go through individual protocols one by one, as we have in prior reviews22, we here discuss the general principle of how expansion microscopy works, seeking a unified workflow picture (Fig. 1a–e). We will not go into detailed protocols in this paper; they are available on the internet23.
First, biomolecules such as proteins8 or RNA9, or labels that bind to biomolecules such as fluorescent antibodies (for proteins)8,5 or fluorescent in situ hybridization (FISH) probes (for RNA)15, are functionalized with chemical handles that allow them to be covalently anchored to the polymer (Fig. 1a, 1b). For example, applying a commercially available small molecule, the succinimidyl ester of 6-((acryloyl)amino)hexanoic acid (acryloyl-X, SE; here abbreviated AcX) will equip amines on proteins (either endogenous proteins, or genetically encoded fluorophores, or applied fluorescent antibodies) with an acrylamide functional group, which can be linked to a growing polyacrylate polymer chain8. Applying a small molecule that contains an alkylating group that reacts to guanine, as well as an acrylamide group (and easily made by mixing two off-the-shelf chemicals), which we call LabelX, enables endogenous RNA (and DNA) to be equipped with a handle that can be linked to a growing polymer chain9; alternatively, applying FISH probes chemically pre-equipped with a similar linker will allow for these probes to be linked to the polymer15.
Next, we synthesize a densely crosslinked sodium polyacrylate mesh throughout the specimen, so that it permeates throughout the cells, between and around the biomolecules and/or labels (Fig. 1b, 1c). We do this by immersing the specimen in a solution containing sodium acrylate monomer (which can form long chains once triggered to polymerize), as well as cross-linking agents so that the final gel topology is a densely linked mesh. The small molecular weight of these building blocks (in the order of magnitute of 100 Daltons) enables their diffusion throughout cells and tissue in a small amount of time, usually less than half an hour for a piece of brain tissue 100 µm thick. This incubation is performed at 4°C, allowing the building blocks to permeate throughout the sample. Polymerization is initiated when the sample is transferred to a 37°C incubator. Within a few hours, a densely cross-linked web of polymer is formed in the sample.
The polymerization process is a standard free radical polymerization process, not unlike the kind used to make polyacrylamide gels for electrophoresis, and similar in spirit to the 1981 protocol of Hausen and Dreyer7 -- a vinyl addition polymerization of sodium acrylate monomers (along with the comonomer acrylamide) and a N-N’-methylenebisacrylamide cross-linker, initiated by the generation of free radicals by a polymerization initiator (e.g., ammonium persulfate (APS)). We add the polymerization accelerator tetramethylethylenediamine (TEMED) as well as a polymerization inhibitor, 4-hydroxy-TEMPO (4-HT), to tune the rate of polymerization so that monomers have time to diffuse throughout the sample before the polymerization reaction takes off. The resultant mesh is extremely dense; small angle x-ray scattering data from similar polymers suggests that the mesh size, or spacing between polymer chains, may be in the 1–2 nanometer range24. Such a small polymer spacing – smaller than the size of many biomolecules themselves -- suggests the possibility of immobilizing biomolecules to the polymer on an individual basis, and also the potential for isotropic expansion (and thus resolution) down to the 1–2 nm range (although that has not yet been experimentally realized).
Next, the specimen – now permeated with swellable polymer, and with key biomolecules or labels bound to the polymer – is mechanically homogenized (Fig. 1c, 1d) so that the components of the tissue do not resist expansion. This can be performed by treatment with high temperatures and detergents8,12, or with proteases that either broadly8,9,14 or specifically8 cleave proteins. In the case of using heat and detergents, the goal is to denature proteins so that they can be easily separated in the expansion step. In the case of using enzymes, the goal is to destroy proteins that are no longer relevant to the later visualization steps, or to chop them up into smaller pieces that can be easily separated. For example, we and others have found that fluorescent antibodies and genetically encoded fluorescent proteins are resistant to proteinase K digestion at doses where most other proteins are chopped up by proteinase K treatment8,14. Thus, for specimens bearing such antibodies or fluorescent proteins, anchoring them to the permeating swellable hydrogel, followed by proteolytic destruction of the other proteins, enables mechanical homogenization while preserving the information to be observed (i.e., the antibody locations or the fluorescent protein locations).
Finally, the addition of water (Fig. 1d, 1e) triggers the swelling of the sodium polyacrylate polymer, so that the biomolecules or labels that are chemically linked to the polymer are pulled apart from each other. Osmotic force draws water into the specimen-polymer composite, and the highly charged carboxyl groups along the polyacrylate backbone then further repel each other (a key advantage of using a polyelectrolyte gel). At that time, labels (e.g., fluorescent antibodies8,12, FISH probes9) can be applied to label biomolecules that were anchored and expanded away from each other, but that are not yet visualizable. Also at that time, amplification of signals can be performed by any of a number of traditional methods, e.g. hybridization chain reaction amplification of FISH signals9, addition of DNA strands equipped with fluorophores10, or use of fluorescent streptavidin to add fluorophores to a previously anchored biotinylated probe21,14. The original expansion microscopy protocol5, as well as many of the follow-on papers8,12,11, reported 4–4.5× linear expansion factors (~100× volumetric expansion) in pure water (Fig. 1f); expanding specimens in a low osmolarity saline solution (helpful to maintain hybridization of post-expansion probes applied to implement FISH) resulted in a ~3.3× linear expansion9.
Through extensive comparison of ExM-expanded samples with pre-expansion samples imaged through traditional super-resolution means (e.g., SIM5,8,11, STORM10), as well as analysis of expanded biomolecular complexes that were previously characterized at a ground-truth nanostructural level (e.g., with electron microscopy)5,10, we were able to show that the expansion process was isotropic, with distortion errors of only a few percent over length scales of tens to hundreds of microns5,8,11. The excellent isotropy arises because of the design of the dense and highly crosslinked swellable polymer, as well as the mechanical homogenization. By analysis of biomolecular complexes of known structure (e.g., microtubules), we further estimated that current expansion processes might be introducing as little as 5–10 nm of error in terms of fundamental resolution10, although pinpointing this number will require further study. As a result, a 4.5× expansion will evenly expand specimens so that when imaged on, say, a confocal microscope with 300 nm lateral resolution, the effective resolution will be 300 / 4.5 ~ 70 nm.
A specimen can be expanded multiple times, for even better effective resolution, a process we call iterative expansion microscopy (iExM)10. A specimen is first expanded as in Fig. 1, except using a chemically cleavable crosslinker rather than N-N’-methylenebisacrylamide to form the initial polyelectrolyte gel network. Such crosslinkers include N,N’-(1,2-Dihydroxyethylene)bis-acrylamide (DHEBA) and N,N’-Bis(acryloyl)cystamine (BAC). Then, after the first expansion is complete, a second swellable polymer (with an uncleavable crosslinker) is synthesized in the space opened up by the first expansion. The biomolecules or labels are transferred from the first gel to the second, the first gel is cleaved, and then the second gel is expanded. This double-expansion process results in a linear expansion factor of about 4.5 × 4.5 ~ 20×, which would result in theory in an effective resolution of 300 nm/20 ~ 15 nm, but in practice is slightly larger because of the size of the labels (i.e., antibodies, linkers) used to stain the specimen in the first place.
In summary, there are different ExM protocols that are optimal for different kinds of specimen and different biological questions. The process of functionalizing the proteins or antibodies with AcX and expanding them away from each other we call protein retention ExM (proExM)8. proExM can be performed by staining the biological samples with primary and secondary antibodies before expansion, or by expanding proteins away from each other before expansion and then adding antibodies afterwards. Other groups developed related strategies for protein retention ExM in parallel to us12,14. The process of functionalizing RNA with LabelX and decrowding RNA molecules in a swellable hydrogel for later FISH visualization we call ExFISH9.
Applications of ExM to neuroscience
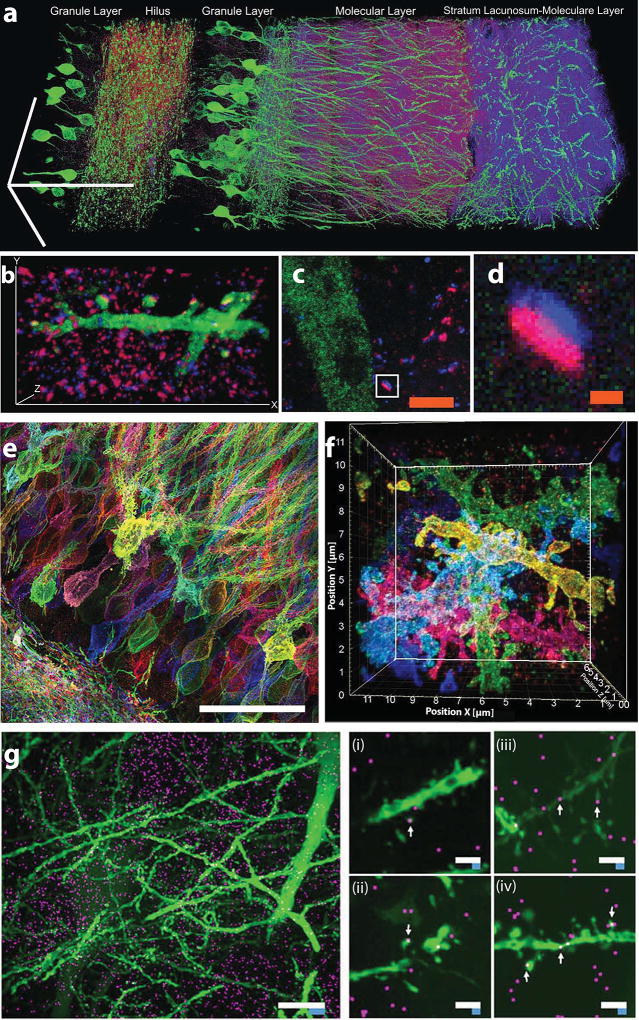
From our earliest paper on ExM, we showed that ExM could be used to visualize synaptic contacts between neurons in brain circuits, e.g. in the mouse hippocampus (Fig. 2a–2d and ref. 5). In particular, with ExM5, proExM8, or iExM10, one can visualize synapses and synaptic proteins (e.g., excitatory and inhibitory neurotransmitter receptors, presynaptic scaffolding proteins, postsynaptic scaffolding proteins, neurotransmitter synthesis enzymes, etc.) in the context of many neurons in a connected circuit, allowing cellular and synaptic analyses to be made across scales in a neural network.
Figure 2. Expansion microscopy of brain circuitry.
a) Expanded mouse hippocampus, with YFP-expressing neurons (green) antibody stained for the postsynaptic protein Homerl (magenta) and the presynaptic protein bassoon (blue) (bars, 100 µm). b) A hippocampal neuron from a piece of mouse brain tissue expanded and labeled as in (a), highlighting a single branch bearing multiple synapses (bars, 13.5 µm ×, 7.3 µm y, 2.8 µm z). c) and d) mouse cortex, expanded and labeled as in (a), with a single synapse (box in c) highlighted in d. (scale bars: c, 2.5 µm; d, 250 nm). Panels a–d adapted from ref. 5. e) Protein retention ExM (proExM, ref. 8) and f) iterative ExM (iExM, ref. 10) of mouse hippocampus expressing Brainbow (i.e., combinatorially expressed fluorophores for randomly labeling neurons with different colors). g) Expansion microscopy fluorescent in situ hybridization (ExFISH, ref. 9) imaging of single RNA molecules (magenta) in mouse hippocampus with simultaneous visualization of protein (green, YFP). Left, Dlg4 mRNA (magenta) visualized simultaneously with YFP (green) (white scale bar, 10 µm; blue scale bar is divided by the expansion factor of 3). Middle (i and ii), dendrites with spine-localized Dlg4 mRNA highlighted with arrows. Right (iii and iv), dendrites with Camk2a mRNA highlighted with arrows (white scale bars, 2µm; blue scale bars are divided by the expansion factor of 3). Panels e–g adapted from the references indicated.
One interesting application of ExM is in the visualization of Brainbow25 (i.e., combinatorially expressed genetically encoded fluorophore)-labeled neural circuitry (via ~70 nm effective resolution proExM in Fig. 2e and ref. 8; via ~25 nm effective resolution iExM in Fig. 2f and ref. 10). Such volumes can be imaged on ordinary confocal microscopes, making neural circuit mapping into a democratized activity requiring hardware of the kind accessible to most groups. A recent algorithmic and experimental study suggests that automated tracing of neural morphologies in expanded brain circuits containing neurons labeled with Brainbow may be possible19.
proExM has successfully revealed synaptic architectures in mouse striatal circuitry20, as well as in the Drosophila brain18 and zebrafish brain26, and has also proven useful in characterizing astrocytic gap junctions near blood vessels in human epilepsy patient brain specimens21, demonstrating its usefulness in a diversity of neuroscientific contexts. proExM has also proven useful in performing a study in the planarian Schmidtea mediterranea that revealed a new non-neural cell type that the authors proposed was a planarian glial cell17.
Mapping of mRNAs and other nucleic acids with subsynaptic precision in intact neural circuits is important for confronting many questions relating to how gene expression is regulated in a spatial fashion, throughout neural circuits, in development, plasticity, and disease. ExFISH has been used to visualize the location and identity of single mRNA molecules, with nanoscale precision, in brain circuits with co-visualization of proteins (Fig. 2g and ref. 9).
Expansion, in addition to providing nanoscale resolution across extended 3-D specimens, enables two other key features. One important observation is that after expansion, the final tissue-gel composite is ~99% water, making the resulting specimens transparent and essentially optical aberration free (Fig. 1g and ref. 5). This makes light sheet imaging into a very fast nanoscale resolution modality, as we have shown by applying lightsheet imaging to expanded samples9 – enabling multiple order-of-magnitude speedup over earlier nanoscale resolution imaging technologies.
A second benefit of ExM is the decrowding of the biomolecules or labels as they are pulled apart during the swelling, which creates room around biomolecules for amplification and analysis chemical reactions. For example, the hybridization chain reaction27 results in many fluorophores being targeted to a single biomolecule via a self-assembling DNA complex. Molecular decrowding makes more room for these large complexes, which otherwise might overlap or compete against each other9. Molecular decrowding may also enable epitopes that may be concealed in protein complexes, to be revealed by separating proteins from one another8. Finally, the decrowding may help, in the future, to support better performance of analytical reactions such as in situ sequencing28, multiplexed antibody staining29, or multiplexed hybridization30, by creating room around biomolecules for well-controlled performance of useful reactions.
Common problems and strategies to overcome them
As with any new technology, early adopters will need to confront potential problems in order to deploy expansion microscopy into their scientific field. This also presents opportunities for refinement and innovation, as various groups have published papers applying or validating expansion microscopy in new contexts like Drosophila18,31, zebrafish26, or human brain21. One common problem that people encounter early in their experiences with expansion microscopy samples regards the fragility of the expanded samples. We recommend storing, transporting, and handling samples as much as possible in the unexpanded state, and expanding in water at the latest feasible moment. Paintbrushes and spatulas can be used to handle gels when unexpanded, but expanded gels require care to transport, for example being carried on coverslips that evenly support the gel22. Storing samples in the compact state may also help preserve fluorescence for longer periods of time than in the expanded state; in an unbuffered solution like pure water, as utilized in the expanded state, fluorophores may deteriorate faster than if samples are stored in phosphate buffered saline (PBS) or another buffered salt solution in the compact state22.
Another common issue is that expanded samples, being volumetrically diluted by a hundredfold or more, can sometimes appear dim when imaged with a microscope, and/or be hard to find since they are so transparent. Amplifying the brightness of labels, as described above, can be very helpful. It is important to validate antibodies in unexpanded samples to insure that they work, since the dilution effect of expansion will compound any antibody errors with additional difficulty of visualization. If antibodies are to be applied post-expansion, it is important not to use proteases that destroy epitopes, but instead to use denaturing conditions to mechanically homogenize cells and tissues32,33. If antibodies are to be applied pre-expansion, it is important not to use cyanine dyes on secondary antibodies, since they will be destroyed during the polymerization process (for a list of alternative dyes that work well in expansion microscopy, see ref. 33).
To facilitate imaging, it can be helpful to trim the gel to the smallest size feasible, and to shape the boundary of the gel (e.g., by cutting the edge in a pattern) so that its shape will tell you its orientation34. It is also very helpful to image samples before expansion and after expansion with a low-magnification microscope to understand where you are in the sample, when imaging a subregion. It can be helpful to immobilize expanded samples in agarose, or to mount expanded samples on a sticky surface, e.g. a polylysine-coated coverslip or slide, to prevent it from drifting during imaging22. For details on how to mount samples for stable imaging in a diversity of microscope settings, and other helpful tips, see ref 22.
Highlights.
Classical super-resolution methods are difficult to use across neural circuits
Expansion microscopy physically magnifies specimens to reveal nanoscale features
Expansion microscopy enables visualization of proteins, RNA, and other biomolecules
Expansion microscopy is easy to use, and rapidly spreading throughout the field
Acknowledgments
For funding E.S.B. acknowledges the HHMI-Simons Fellowship, John Doerr, the Open Philanthropy project, IARPA D16PC00008, NIH grants 1R01MH103910, 1RM1HG008525, 1R01MH110932, 1R01EB024261 and 1R01NS102727, the Cancer Research UK Grand Challenge, and U. S. Army Research Laboratory and the U. S. Army Research Office under contract/grant number W911NF1510548. We thank all members of the Synthetic Neurobiology group for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, Roberts M, Morgan JL, Tapia JC, Seung HS, Roncal WG, Vogelstein JT, Burns R, Sussman DL, Priebe CE, Pfister H, Lichtman JW. Saturated Reconstruction of a Volume of Neocortex. Cell. 2015;162(3):648–661. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Eichler K, Li F, Litwin-Kumar A, Park Y, Andrade I, Schneider-Mizell CM, Saumweber T, Huser A, Eschbach C, Gerber B, Fetter RD, Truman JW, Priebe CE, Abbott LF, Thum AS, Zlatic M, Cardona A. The complete connectome of a learning and memory centre in an insect brain. Nature. 2017;548(7666):175–182. doi: 10.1038/nature23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He J, Zhou R, Wu Z, Carrasco MA, Kurshan PT, Farley JE, Simon DJ, Wang G, Han B, Hao J, Heller E, Freeman MR, Shen K, Maniatis T, Tessier-Lavigne M, Zhuang X. Prevalent presence of periodic actin–spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc Natl Acad Sci. 2016;113(21):6029–6034. doi: 10.1073/pnas.1605707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Este E, Kamin D, Göttfert F, El-Hady A, Hell SW. STED Nanoscopy Reveals the Ubiquity of Subcortical Cytoskeleton Periodicity in Living Neurons. Cell Rep. 2015;10(8):1246–1251. doi: 10.1016/j.celrep.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 5••.Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science (80-) 2015;347(6221):543–548. doi: 10.1126/science.1260088. The paper introducing the concept, demonstration, and initial validation of expansion microscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka T, Fillmore D, Sun ST, Nishio I, Swislow G, Shah A. Phase Transitions in Ionic Gels. Phys Rev Lett. 1980;45:1636–1639. doi: 10.1103/PhysRevLett.45.1636. [DOI] [Google Scholar]
- 7.Hausen P, Dreyer C. The use of polyacrylamide as an embedding medium for immunohistochemical studies of embryonic tissues. Stain Technol. 1981;56(5):287–293. doi: 10.3109/10520298109067329. [DOI] [PubMed] [Google Scholar]
- 8•.Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu C-C (Jay), English BP, Gao L, Martorell A, Suk H-J, Yoshida F, Ellen M, DeGennaro DHR, Gong G, Seneviratne U, Tannenbaum SR, Desimone R, Cai D, Boyden ES. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34:987–992. doi: 10.1038/nbt.3625. A practical method for performing expansion microscopy for protein visualization, using commercially available chemicals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Shoh Asano ERD, Chang J-B, Marblestone A, Church GM, Raj A, Boyden ES. Nanoscale imaging of RNA with expansion microscopy. Nat Methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. A practical method for performing expansion microscopy for RNA visualization, using commercially available chemicals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Chang J-B, Chen F, Yoon Y-G, Jung EE, Babcock H, Kang JS, Asano S, Suk H-J, Pak N, Tillberg PW, Wassie AT, Cai D, Boyden ES. Iterative expansion microscopy. Nat Methods. 2017 Apr; doi: 10.1038/nmeth.4261. By expanding a sample over and over, one can achieve higher magnifications than possible with a single step expansion protocol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Bucur O, Irshad H, Chen F, Weins A, Stancu AL, Oh E-Y, DiStasio M, Torous V, Glass B, Stillman IE, Schnitt SJ, Beck AH, Boyden ES. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat Biotechnol. 2017 doi: 10.1038/nbt.3892. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Ku T, Swaney J, Park J-Y, Albanese A, Murray E, Cho JH, Park Y-G, Mangena V, Chen J, Chung K. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol. 2016 doi: 10.1038/nbt.3641. In press(November 2015) doi:10.1038/nbt.3641. Another paper describing how to perform expansion microscopy for protein visualization, using commercially available chemicals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang YS, Chang J-B, Alvarez MM, Trujillo-de Santiago G, Aleman J, Batzaya B, Krishnadoss V, Ramanujam AA, Kazemzadeh-Narbat M, Chen F, Tillberg PW, Dokmeci MR, Boyden ES, Khademhosseini A. Hybrid Microscopy: Enabling Inexpensive High-Performance Imaging through Combined Physical and Optical Magnifications. Sci Rep. 2016;6:22691. doi: 10.1038/srep22691. (November 2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Chozinski TJ, Halpern AR, Okawa H, Kim H-J, Tremel GJ, Wong ROL, Vaughan JC. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods. 2016 Apr;13:1–7. doi: 10.1038/nmeth.3833. Another paper describing how to perform expansion microscopy for protein visualization, using commercially available chemicals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsanov N, Samacoits A, Chouaib R, Traboulsi A-M, Gostan T, Weber C, Zimmer C, Zibara K, Walter T, Peter M, Bertrand E, Mueller F. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw784. gkw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A, Kim JH, Ranjan P, Metcalfe MG, Cao W, Mishina M, Gangappa S, Guo Z, Boyden ES, Zaki S, York I, García-Sastre A, Shaw M, Sambhara S. Influenza virus exploits tunneling nanotubes for cell-to-cell spread. Sci Rep. 2017 Jan;7:40360. doi: 10.1038/srep40360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang IE, Lapan SW, Scimone ML, Clandinin TR, Reddien PW. Hedgehog signaling regulates gene expression in planarian glia. Elife. 2016;5 doi: 10.7554/eLife.16996. (September 2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosca TJ, Luginbuhl DJ, Wang IE, Luo L. Presynaptic LRP4 promotes synapse number and function of excitatory CNS neurons. Elife. 2017;6 doi: 10.7554/eLife.27347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sümbül U, Roossien D, Chen F, Barry N, Boyden E, Cai D, Cunningham JP, Paninski L. Automated scalable segmentation of neurons from multispectral images. Adv Neural Inf Process Syst. 2016 [Google Scholar]
- 20.Crittenden JR, Tillberg PW, Riad MH, Shima Y, Gerfen CR, Curry J, Housman DE, Nelson SB, Boyden ES, Graybiel AM. Striosome-dendron bouquets highlight a unique striatonigral circuit targeting dopamine-containing neurons. Proc Natl Acad Sci U S A. 2016;113(40):11318–11323. doi: 10.1073/pnas.1613337113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Deshpande T, Li T, Herde MK, Becker A, Vatter H, Schwarz MK, Henneberger C, Steinhäuser C, Bedner P. Subcellular reorganization and altered phosphorylation of the astrocytic gap junction protein connexin43 in human and experimental temporal lobe epilepsy. Glia. 2017 doi: 10.1002/glia.23196. A study which applied expansion microscopy to human brain tissue in a clinical context. [DOI] [PubMed] [Google Scholar]
- 22•.Gao R, Asano SM, Boyden ES. Q&A: Expansion microscopy. BMC Biol. 2017;15(1):50. doi: 10.1186/s12915-017-0393-3. Helpful tips on how to do expansion microscopy in a variety of laboratory contexts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ExpansionMicroscopy.org: Physical Specimen Expansion Enabling 3-D Large Volume, Nanoscale Imaging [Internet] Available from: http://expansionmicroscopy.org/
- 24.Cohen Y, Ramon O, Kopelman IJ, Mizrahi S. Characterization of inhomogeneous polyacrylamide hydrogels. J Polym Sci Part B Polym Phys. 1992;30:1055–1067. doi: 10.1002/polb.1992.090300913. [DOI] [Google Scholar]
- 25.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10(6):540–7. doi: 10.1038/nmeth.2450. [DOI] [PubMed] [Google Scholar]
- 26.Freifeld L, Odstrcil I, Förster D, Ramirez A, Gagnon JA, Randlett O, Costa EK, Asano S, Celiker OT, Gao R, Martin-Alarcon DA, Reginato P, Dick C, Chen L, Schoppik D, Engert F, Boyden ES. Expansion Microscopy of Zebrafish for Neuroscience and Developmental Biology Studies. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1706281114. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi HMT, Beck Va, Pierce Na. Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano. 2014;8(5):4284–4294. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SSF, Li C, Amamoto R, Peters DT, Turczyk BM, Marblestone AH, Inverso SA, Bernard A, Mali P, Rios X, Aach J, Church GM. Highly Multiplexed Subcellular RNA Sequencing in Situ. Science (80-) 2014 Mar;343(6177):1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jungmann R, Avendaño MS, Woehrstein JB, Dai M, Shih WM, Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014;11(3):313–8. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, Zhuang X. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci. 2016 Dec;113(50):14456–14461. doi: 10.1073/pnas.1617699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cahoon CK, Yu Z, Wang Y, Guo F, Unruh JR, Slaughter BD, Hawley RS. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc Natl Acad Sci. 2017:201705623. doi: 10.1073/pnas.1705623114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, Choi H, Park YG, Park JY, Hubbert A, McCue M, Vassallo S, Bakh N, Frosch MP, Wedeen VJ, Seung HS, Chung K. Simple, Scalable Proteomic Imaging for High-Dimensional Profiling of Intact Systems. Cell. 2015;163(6):1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu C-C, English BP, Gao L, Martorell A, Suk H-J, Yoshida F, DeGennaro EM, Roossien DH, Gong G, Seneviratne U, Tannenbaum SR, Desimone R, Cai D, Boyden ES. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016;34(9):987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.proExM for tissues: gelation demonstration [Internet] Available from: https://www.youtube.com/watch?v=OksNCAJwxVI.