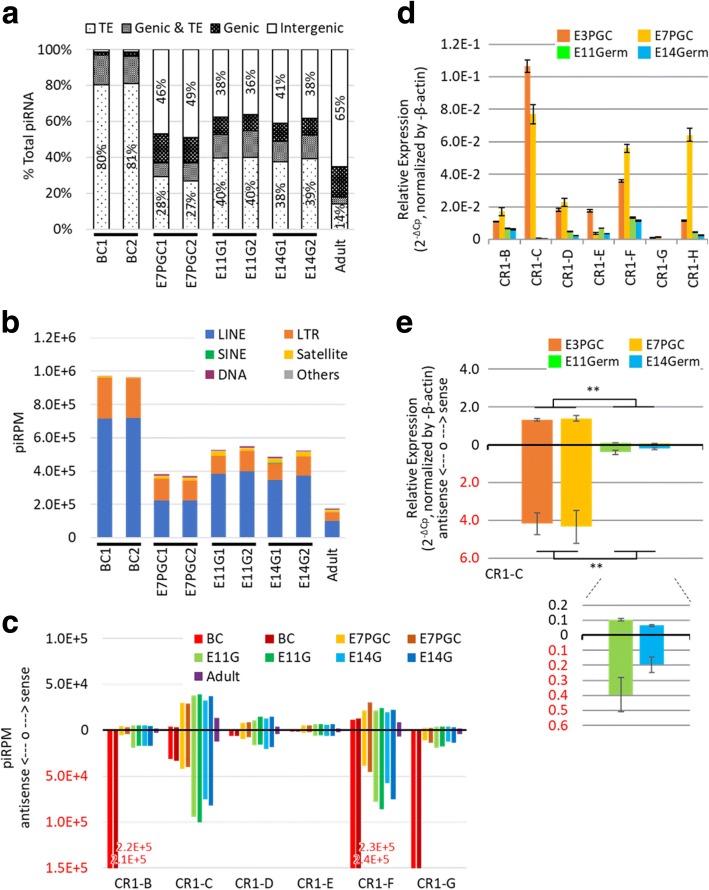
Fig. 2.
Developmental stage-dependent genomic associations and TE expression modulation of piRNA candidates. a Genomic association varied in stage-dependent fashion. Each feature was calculated in proportion to the respective total piRNA candidates. b PiRNA candidates mapped to TEs. piRPM is calculated for each enlisted category. c The stage-dependent association of piRNAs to subfamilies of LINEs. PiRNA sequences are preferentially mapped antisense to TEs. d Expression of CR1 subfamily members from enriched embryonic germ cells. e Stranded RT-qPCR analysis (N = 3) over CR1-C transcription among cultured E3 circulating PGC (E3PGC), cultured E7 gonadal PGC (E7PGC), and germ cell enriched population, E11Germ and E14Germ, from E11 and E14 Gonads, respectively. ** represents p-value < 0.01