Fig. 3.
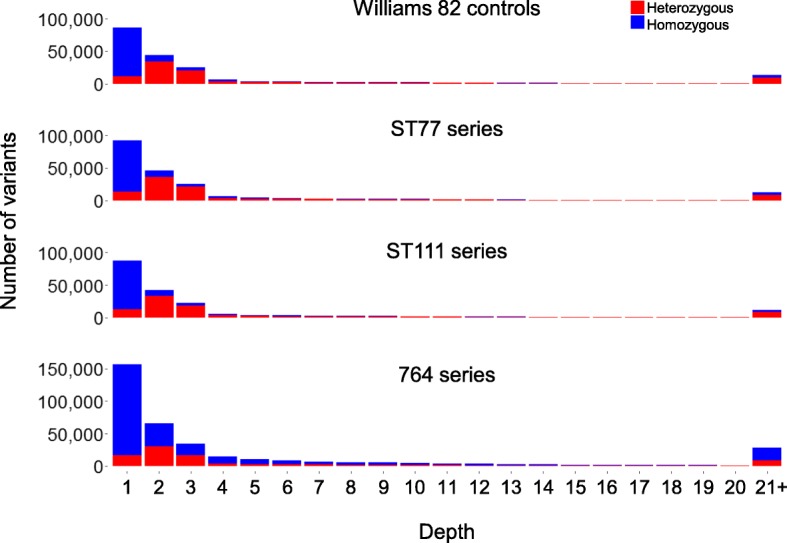
Depth of sequence coverage for all polymorphic variants (SNPs and indels) called in the Lambirth et al. [22] study. The polymorphic calls shown here were made between each sample and the reference genome ‘Williams 82’, without consideration for the uniqueness of the call among series or reproducibility among different plants within the series. Homozygous calls are shown in blue and heterozygous calls are shown in red. Each bar sums the number of polymorphisms across the nine plants that were called at each read depth (e.g., we are showing the ~ 211,448 total variants called in series ST77 across the nine plants; ST77 averaged 23,494 variants per plant). Note the relatively larger peak in the 21+ category for the 764 series compared to the other series; many of these (mostly homozygous) calls likely represent standing variants between lines ‘Thorne’ and ‘Williams 82’. The 21+ peaks in the other three groups (ST77, ST111, and ‘Williams 82’ controls) may derive from various factors, most obviously the clusters of variants that are found within heterogeneous regions of different sub-lines of ‘Williams 82’
