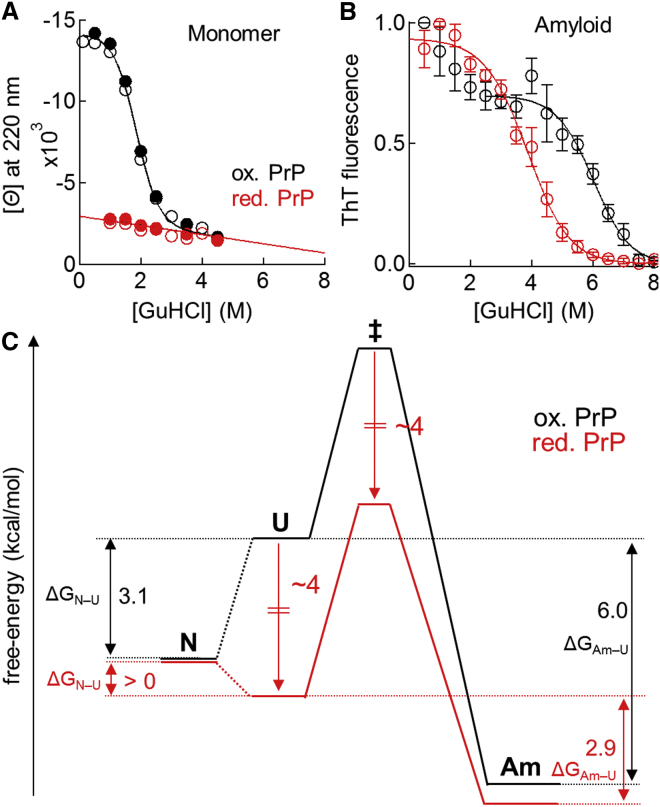
Figure 4.
(A) GuHCl-unfolding curves for the monomeric forms of oxidized (black circles) and reduced PrP (red circles) at the protein concentration of 10 (open circles) or 25 μM (solid circles). (B) GuHCl-unfolding curves for amyloid fibrils generated from oxidized (black open circles) and reduced PrP (red open circles). Data represent mean values ± standard deviations from three experiments that used different preparations of amyloid fibrils. (C) Effects of disulfide bond reduction on the free-energy landscape of protein misfolding at a protein concentration of 2.5 μM in a denaturant-free solution. Disulfide bond reduction decreased the free-energies of the unfolded (U) and transition states (‡) by ∼3–4 kcal/mol, without significantly changing the free-energies of the native state (N) and amyloid fibrils (Am), leading to net destabilization effects of >3.1 kcal/ mol on the N state (Fig. 4A), 3.1 kcal/mol on the Am state (Fig. 4B), but ∼0 kcal/mol on the ‡ state when compared to the U state (Fig. 5B). To see this figure in color, go online.