Figure 3.
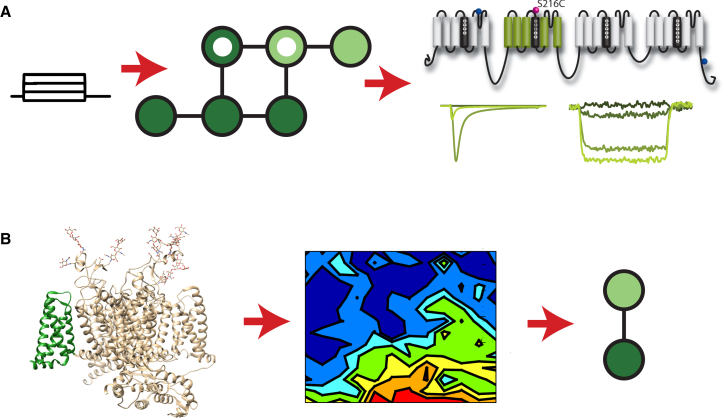
A path to molecularly detailed kinetic models of complex channels. The complexity of four-domain channels complicates the approach used for K+ channels in Figs. 1 and 2. An alternative approach is to use VCF experiments in combination with state-mutating algorithms to create models that represent only a single VSD. VCF data in the figure are adapted from (34). (A) Kinetic models of ion channels are typically parameterized to recapitulate ionic-current dynamics that result from a series of pulse protocols. One or more states in the model graph are identified as open (inner white circles). By also identifying states as having a resting (darker circles) or active (lighter circles) VSD, fluorescence dynamics can also be simulated, allowing a single VSD in a complex channel to be explicitly represented. Shown are the fluorescence-reporting conformation of domain II of the cardiac Na+ channel and the ionic current that was recorded simultaneously. (B) Once the relationship between the VSD transition and the rest of the channel is modeled, an energy landscape derived from MD simulations can be used to simulate the VSD transition. The structure shown was rendered from PDB: 5X0M (26) of the eukaryotic cockroach Na+ channel with the domain II VSD shown in green. This approach reduces the need to derive models from simulations of the whole channel, which are not currently tenable.