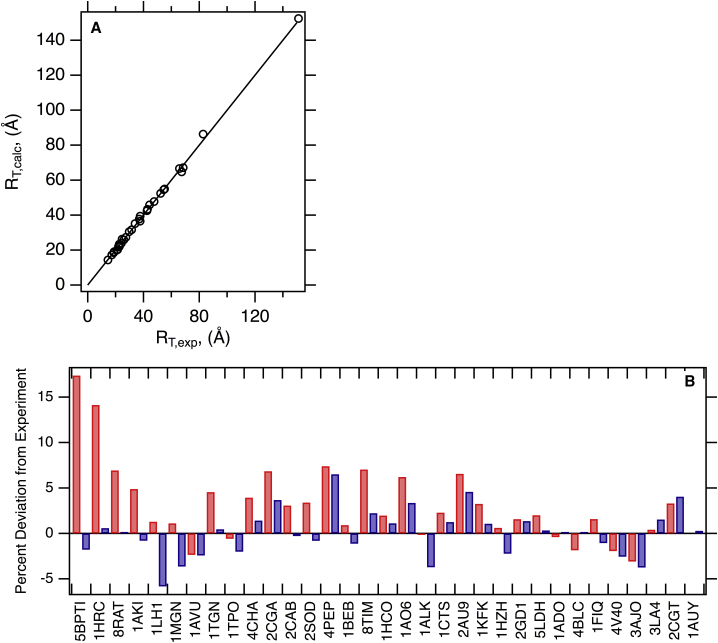
Figure 3.
The convex hull method accurately predicts experimental protein translational hydrodynamic radii. (A) HullRad predicted RT,calc for folded proteins are plotted versus experimental RT,exp values. The black line represents a slope of one and intercept of zero; the correlation coefficient for a linear regression of the data (data not shown) is 0.998. (B) Percent signed errors of calculated to experimental RT for folded proteins are plotted as bars. Dark gray (blue online) was calculated with HullRad; light gray (red online) was calculated with HYDROPRO. In this comparison, all HYDROPRO calculations used atomic-level beads except the structure 2CGT, for which residue level beads were used. Only HullRad was used to calculate hydrodynamic properties for the largest example, 12AUY. The individual proteins are labeled according to respective PDB IDs; common names, molecular masses, number of chains, and respective hydrodynamic radii are listed in Table 1. This figure was created in Igor. To see this figure in color, go online.