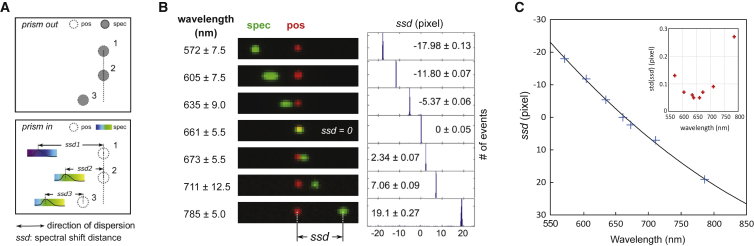
Figure 2.
Calibration of the single-molecule spectral imaging system. (A) Principles of spectral measurement on the single-molecule spectral imaging system are shown. The positional (empty circle) and spectral (filled circle) images were first aligned with high precision before the prism was inserted (top panel). With the prism inserted, the spectral images of all fluorescent objects became elongated, and the intensity profile of each elongated image represented the emission spectrum of the corresponding object (bottom panel). For each object in the overlaid image, its ssd was defined as the distance between the centroid of its positional image and the (sub-) pixel position of maximum emission intensity in the spectral image. In this case, objects two and three are the same type of fluorophore and they exhibit the same ssd (i.e., ssd2 = ssd3) values, whereas object one is in a different color exhibiting a different ssd value (ssd1); (B) Overlaid positional (pos) (red) and spectral (spec) (green) images of fluorescent beads (broad emission between 500 and 800 nm when excited at 488 or 561 nm) after passing through a series of narrow bandpass filters as indicated. The data point at ∼572 nm was taken with 488 nm excitation and the remaining data were taken with 561 nm excitation. The positional and spectral images were intentionally overlapped with subpixel precision at 661 ± 5.5 nm (i.e., ssd = 0.0). Thus, images acquired at shorter wavelengths had negative ssd with the spectral image to the left of the positional image, and the opposite in images taken at longer wavelengths (left). Shown on the right, histograms of ssd at each wavelength with the means and SD indicated. (C) The calibration curve showing the relationship between the center wavelength and the measured ssd values is shown. Inset is the SD of ssd at each wavelength. To see this figure in color, go online.