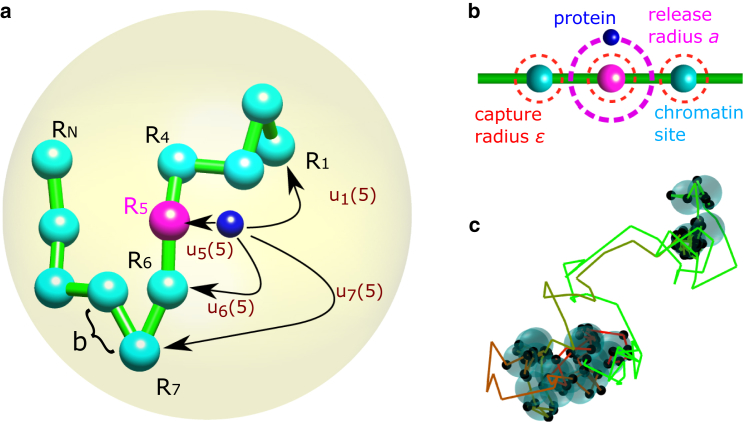
Figure 1.
Transition between monomer sites. (a) A particle (dark blue) representing a protein interacts with a monomer site (cyan) that serves as a chromatin locus. It detaches from an initial monomer n (magenta) and reattaches to site l with probability without touching other sites along the way. The polymer is inside a confining domain (yellow) of radius A. b is the mean-square displacement of a bond. (b) The particle is released at distance a from the initial site, with uniform angular distribution. It attaches to a site once it is at distance ϵ from it. We depict chromatin as a coarse-grained chain of beads. Each bead is of characteristic size b, representing 3.2 kb and of size 30 nm (41). For the rest of the paper, the characteristic length in our system is b. Hence, for and nm, the release and capture radii are of the order of the size of a protein and the interaction distance. (c) Trajectory of the particle interacting with the monomer sites (cyan). The trajectory color changes with time from red to green. The black dots are attachment points at the monomer site (,, , and ). To see this figure in color, go online.