Figure 3.
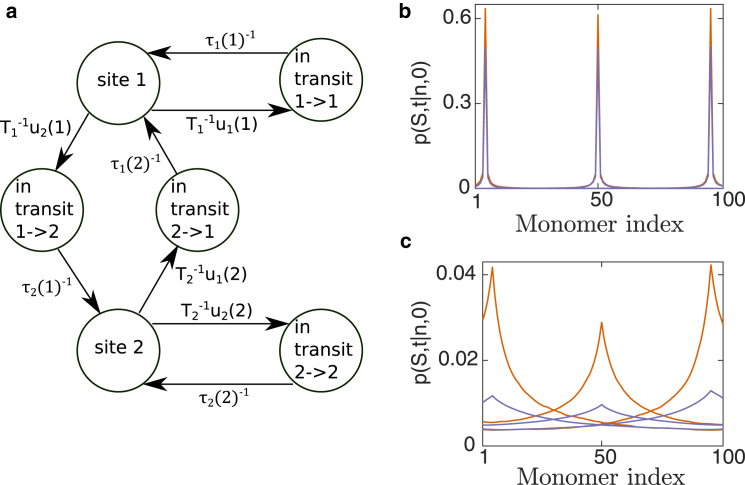
Site-site transition time and particle distribution in the domain. (a) Formulating the behavior of a particle (protein) that interacts with the polymer (chromatin), inside a domain (nucleus) as a continuous-time Markov chain. We illustrate it for the case of two binding monomer sites. The particle can be bound at site 1 or 2 and is released with Poissonian dissociation rates and. While diffusing to bind to a site, it is “in transit” and arrives at its destination with probability 1 and rate . (b and c) The monomer is bound at a site for a characteristic time The probability, p, of the particle being in a bound state at the monomer site after 1T (b) or 10T (c) is computed by solving numerically Eq. 6. The monomer starts at a distance a from either monomer 2, 50, or 99. and were estimated from Brownian simulation: , , , and (orange) or (purple). For , there are 100 bound states and in-transit states. To see this figure in color, go online.