Abstract
Force plays a key role in regulating dynamics of biomolecular structure and interactions, yet techniques are lacking to manipulate and continuously read out this response with high throughput. We present an enzymatic assay for force-dependent accessibility of structure that makes use of a wireless mini-radio centrifuge force microscope to provide a real-time readout of kinetics. The microscope is designed for ease of use, fits in a standard centrifuge bucket, and offers high-throughput, video-rate readout of individual proteolytic cleavage events. Proteolysis measurements on thousands of tethered collagen molecules show a load-enhanced trypsin sensitivity, indicating destabilization of the triple helix.
Introduction
The conventional view of fixed protein structure is evolving toward one of dynamic conformational sampling. This change in perspective requires techniques to interrogate structures that are not static. Although the structural technique of NMR spectroscopy offers atomic-level resolution of time-averaged dynamics of small proteins in solution, it lacks the ability to resolve larger structures and complexes. For larger systems, transient accessibility of otherwise buried regions can be identified and quantified using enzymatic cleavage assays (1, 2, 3). These assays provide significant insight into structural stability, but because they require electrophoretic analysis of products extracted at discrete time points, the assays lack real-time monitoring of reaction progress. Furthermore, none of these assays assesses the modification of structures by applied force. Force is an increasingly apparent regulator of cellular activity, wherein mechanical actuation of cryptic sites in proteins can control downstream signaling (4). There is a need for new methods capable of reading out, in real time, changes in protein conformation resulting from external stimuli such as applied force.
Here, we introduce the technique of high-throughput, force-dependent proteolysis using a mini-radio centrifuge force microscope (MR.CFM), which enables the real-time assessment of molecular structural stability. As proof of concept, we assess the force-dependent modulation of collagen’s triple helical structure.
Collagen is a key component of the extracellular matrix and is the predominant protein in vertebrates, where it confers tensile strength to connective tissues (5, 6). Collagen is a hierarchically structured material: individual triple-helical collagen proteins assemble into highly ordered fibrils, which in turn are the building blocks for fibers, a wide range of connective tissues, and the extracellular matrix. How these are regulated by applied load, in particular their susceptibility to enzymatic remodeling, has direct relevance to their physiological performance. Numerous studies have reached contradictory conclusions about how applied force influences the proteolytic susceptibility of collagen at different hierarchical scales, and a clear mechanistic interpretation of the varied results remains elusive (6, 7). Understanding how collagen is affected by stress is complicated by the embedded hierarchical structural levels and their respective responses to force.
At the molecular level, collagen is distinguished by its unique triple-helical structure. This structure prevents conventional proteolysis and requires specialized collagenases for its degradation (8). Not surprisingly, given its mechanical importance, there has been substantial interest in understanding how the triple helix deforms when stretched. However, previous studies have reached contradictory conclusions about how the triple helix is modified by applied force (Table 1): does it entropically extend, shear open, unwind, or tighten? These investigations each have limitations: model peptide sequences lack the context of the full-length protein (6, 9); triple-helical stability can depend on the force field used in simulations (6, 10, 11); changes in length of the full protein observed under force do not reveal structural or sequence-dependent information (12, 13); and collagenases used to investigate force-induced deformations (9, 14, 15) are not inert probes, because their specialized proteolysis of intact collagen derives from their ability to manipulate its triple-helical structure (8, 16).
Table 1.
Reported Force-Dependent Responses of Collagen’s Triple Helix for Forces below 10 pN
| Force Response | Technique Used |
|---|---|
| Stabilizes/tightens | force-dependent cleavage with bacterial collagenase (14) |
| steered molecular dynamics (6) | |
| No change (entropically extends) | optical tweezers stretching (12) |
| steered molecular dynamics (6) | |
| force-dependent cleavage with bacterial collagenase (15) | |
| Destabilizes/unwinds/lengthens | force-dependent cleavage with MMP1 (9, 15) |
| optical tweezers stretching (13) | |
| Shears open | steered molecular dynamics (10) |
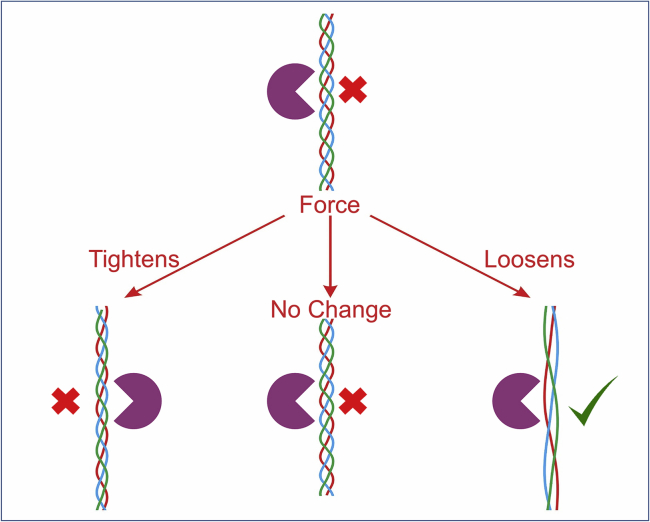
To resolve the question of how collagen’s structure is altered by force, we implement the hallmark assay for triple-helical stability, trypsin susceptibility (1), enhancing its utility by incorporating force into single-molecule proteolysis assays. A tight triple helix blocks proteolytic cleavage by presenting a steric barrier that prevents access of a single polypeptide chain to trypsin’s active site. Thus, only if collagen’s structure locally denatures can cleavage by trypsin occur (1). If a stretching force destabilizes the triple helix, an increase in the proteolysis rate should be observed (Fig. 1). Conversely, if no change in structure occurs, or if the helix tightens, the cleavage rate should remain unchanged or be reduced. To assess the effect of force on collagen’s stability, we use MR.CFM to probe thousands of individual collagen molecules and determine how force affects their rate of proteolysis by trypsin.
Figure 1.
Possible responses of collagen’s triple helix to applied stress. In the absence of applied force, a stable triple helix is resistant to proteolysis by trypsin (upper). If the helix tightens or remains unchanged by force (lower left and center, respectively), it remains resistant to cleavage by trypsin. Only if force induces a destabilization of the helix will proteolysis by trypsin be possible (lower right). To see this figure in color, go online.
Materials and Methods
MR.CFM
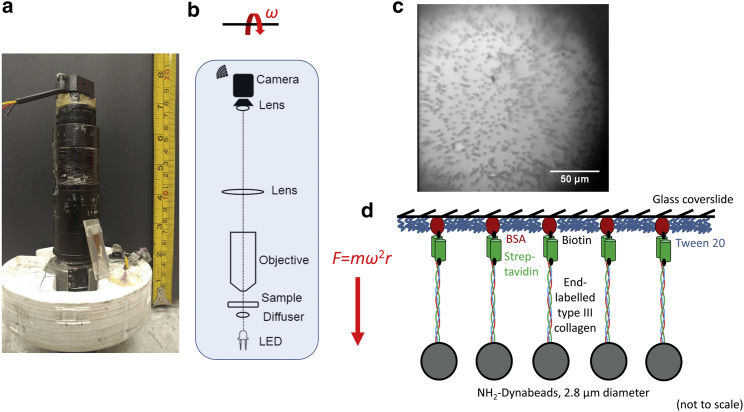
MR.CFM is a wireless, fully self-contained, compact bright-field microscope (Fig. 2) that exploits the rotation of a commercial benchtop centrifuge to exert controlled force. MR.CFM fits within a single bucket of a Beckman Coulter (Brea, CA) Allegra X-12R centrifuge, equipped with an SX4750A ARIES Swinging Bucket Rotor assembly. This assembly has an imbalance tolerance of ±50 g. MR.CFM was built using as many commercial parts as possible for ease of reproduction. The final height and weight for MR.CFM are 154 mm and 421 g (including batteries), respectively. These fall well within the length and weight budgets of the centrifuge rotor.
Figure 2.
Mini-radio centrifuge force microscope (MR.CFM) and collagen tethering. (a) Photograph of the microscope, with the centrifuge bucket insert at the bottom and a sample chamber leaning at the side. The height of the assembly is <16 cm and its mass is 421 g. (b) Schematic of the optical elements within the microscope. A force F = mω2r is exerted on all beads of relative mass m within the sample chamber, located a distance r from the central rotation axis, when the rotor spins at angular frequency ω. (c) Image of microspheres subjected to 9 pN of force in MR.CFM, tethered to the surface by collagen molecules. Although each bead occupies only a few pixels in the final image, they are easily detected. (d) Schematic of tethering geometry. Single molecules of collagen are tethered to a glass slide via biotin-streptavidin linkages, and are covalently linked to the surface of heavy beads via thiol-amine coupling. To see this figure in color, go online.
Schematics and parts lists are provided in Fig. S1 and Table S1, with a description of the design provided in the Supporting Materials and Methods. Drawings of the few custom parts are provided in Fig. S2 and in computer-aided drawings in Data S1. Note that although the designs provided are specific to this rotor assembly, MR.CFM can easily be adapted for other benchtop centrifuges, primarily by modifying the base.
Image acquisition and particle detection
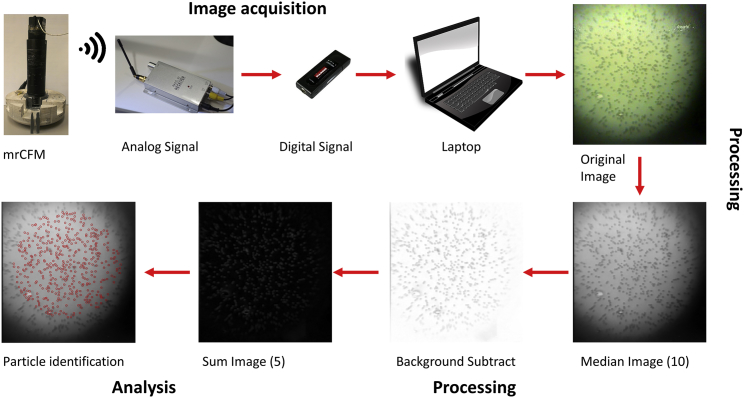
The image-analysis workflow to acquire, process, and analyze data from the raw image stream is shown schematically in Fig. 3 and is described in the Supporting Materials and Methods. The ImageJ macro written for image processing is provided as Data S2. Particles are counted with the Mosaic particle tracker 2D/3D plugin for ImageJ (17).
Figure 3.
Work flow for MR.CFM acquisition, processing, and analysis of bead images. Images are acquired with the wireless camera in MR.CFM, then transmitted via radio signal to the audiovisual receiver. The audiovisual receiver collects the analog signal, which is output to a USB analog-to-digital converter attached to a laptop computer, where the image stream is simultaneously visualized in real time and stored. The original output image files from MR.CFM (shown here at 9 pN of force) are processed in ImageJ to improve contrast and remove rotation-speed-dependent interference. Particles are counted using the Mosaic particle-tracking program. The resultant overlaid image visually shows that this procedure is robust and identifies beads within the defined search area. To see this figure in color, go online.
MR.CFM sample chambers
Glass slides and coverslips were cut to 12 mm × 33 mm to fit the sample holder. Sample chambers were created between a coverslip (borosilicate glass no. 1; VWR, 48393 106) and a glass slide (soda lime glass 2 mm, Logitech Limited, Newark, CA) by strips of glue (JB Weld, Atlanta, GA), with a depth of ∼300 μm.
Surface chemistry
We implemented a blocking and specific tethering system that relies on a self-assembled monolayer of Tween-20 for blocking and on biotinylated bovine serum albumin (BSA) for specific tethering. Modifications to the published protocol for surface blocking (18) are provided in the Supporting Materials and Methods.
Collagen functionalization
Type III collagen was used in these experiments due to its chemically accessible C-terminal cysteine residues, unique among fibrillar collagens. Recombinant human type III collagen (Fibrogen FG-5016) was end-labeled for manipulation as previously described (19), with information provided in the Supporting Materials and Methods. Briefly, this approach utilized biotinylation of introduced N-terminal aldehydes and covalent linking of C-terminal thiols to amine-functionalized superparamagnetic microspheres (2.8 μm diameter, M270 Dynabeads).
Single-molecule proteolysis experiments
All trypsin cleavage experiments were performed at room temperature with the following final concentrations: trypsin (T1426; Sigma Aldrich, St. Louis, MO) 2.0 mg/mL, 1× reaction buffer (0.1 M Tris and 0.4 M NaCl, pH 7.4), and collagenated beads (<108 beads/mL). To minimize room-temperature proteolysis of collagen by trypsin (Fig. S3), all solutions were kept at 4°C before use. Control experiments on DNA used aldehyde- and thiol-functionalized DNA (see Supporting Materials and Methods). Experiments at 9 pN of force were performed in MR.CFM, whereas zero-force (67 fN) experiments utilized a bright-field microscope (BX51; Olympus, Tokyo, Japan, equipped with 10× Plan N objective). Detailed experimental protocols are provided in the Supporting Materials and Methods, as is information about the number of distinct experimental runs for each condition.
The number of potential trypsin cleavage sites in type III collagen was given by the Expasy PeptideCutter tool (20), querying from sites 154 to 1221 of UniProtKB: P02461. This sequence represents the collagen domain of human type III procollagen.
Data analysis
Fitting of the decay data with (2a), (2b) was performed in IGOR Pro (WaveMetrics, Lake Oswego, OR), weighted by the errors associated with each point. Data were analyzed following three separate approaches: determining from all replicates the mean fraction remaining as a function of time (where the error used was the standard error of the mean) (Fig. 4; Table 2); pooling all replicates to analyze the fraction of the total beads remaining as a function of time (where the error was taken to be a counting error) (Table S2 for collagen and Fig. S4 for DNA); and by fitting each individual replicate to Eq. 1 (where the error at each time point was taken to be a counting error) and determining the mean of these rates (Table S3 for collagen and Table S4 for DNA). Fit parameters are presented as best-fit values ± SD. To determine whether force enhances or hinders collagen proteolysis, we compared the trypsin-dependent rates kTr,F and kTr,0.
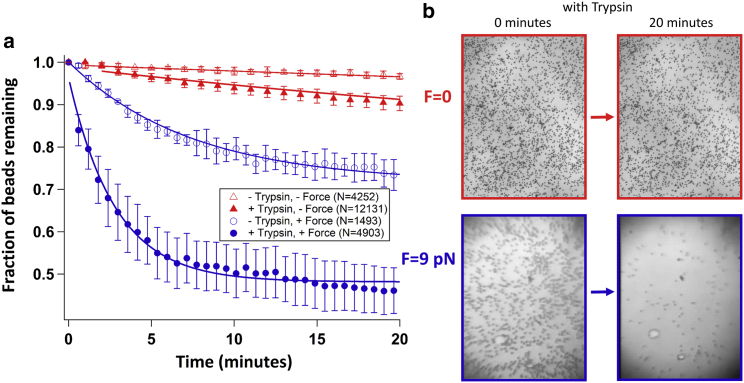
Figure 4.
Collagen proteolysis by trypsin is enhanced by force. (a) Collagen tethers rupture at higher rates in the presence of force and trypsin (solid blue circles) than in the absence of either, implying their force-induced denaturation. Data points represent the mean fraction of beads remaining at each time and error bars represent standard errors of this mean. Every 10th data point is shown for the experiments at 9 pN of force, for clarity. Each experimental condition is well described by a single-exponential decay process, with rate constants presented in Table 2. (b) Representative images of collagen-tethered beads at the beginning (left) and after 20 min (right) of incubation with trypsin. Top: no force (67 fN; gravitational force on beads), recorded using a conventional bright-field microscope. Bottom: 8.8 pN of force, recorded within MR.CFM. To see this figure in color, go online.
Table 2.
Rate Constants Obtained for Collagen Bead Detachment
| Experiment | keff (min−1) | kns (min−1) | kTr (min−1) |
|---|---|---|---|
| − trypsin, F = 0 | – | 0.010a ± 0.013 | – |
| + trypsin, F = 0 | 0.020 ± 0.001 | 0.010b ± 0.013 | 0.009 ± 0.013 |
| − trypsin, F = 9 pN | – | 0.135 ± 0.003 | – |
| + trypsin, F = 9 pN | 0.357 ± 0.018 | 0.135b ± 0.003 | 0.222 ± 0.018 |
Rate constants were determined by weighting <f(t)> by the mean ± SE.
Determined by weighting f(t) by counting error (Table S2), as the mean ± SE-based kns = 0.007 min−1 carried a very high uncertainty.
Results and Discussion
MR.CFM
To perform high-throughput proteolysis assays in a force-dependent manner, we developed an instrument dubbed the MR.CFM (Fig. 2). Centrifuge force microscopy requires only a microscope capable of stable imaging at high acceleration and dense particles on which to exert force, and can have a force range orders of magnitude larger than that of magnetic or optical tweezers (21). Our instrument was designed to be easily adopted by biological research labs, as it is a “plug-and-play,” modular, entirely wireless brightfield microscope that can be placed into the bucket of a conventional benchtop centrifuge, counterbalanced simply by tubes of water. The force is determined by the rotational frequency of the centrifuge, ω (typically hundreds to thousands of rotations per minute), the distance between the sample and the axis of rotation, r (typically tens of centimeters), and the mass of the bead, m (relative to the displaced water), tethered to the molecule of interest: F = mω2r. Unlike the limited region of constant force in alternative single-molecule force techniques (22), the CFM provides a controllable, uniform force across the entire rotating sample chamber.
The layout of MR.CFM was driven by optical simplicity and low mass. Thus, in contrast to alternative approaches (23, 24), our instrument has a straight optical path and requires no mirrors. Besides ease of optical design, the straight layout positions the camera closer to the center of rotation, subjecting it to less force during operation. In addition to substantially reduced cost, MR.CFM affords significant advantages over previous compact designs by 1) utilizing an unmodified centrifuge, thereby being amenable to use in shared facilities (24); and 2) offering continuous radio-frequency video output, permitting unlimited video-rate capture and digitization (23). The latter advantage confers the ability to read out molecular kinetics over experimental timescales of hours, without the need to sacrifice temporal resolution.
With a CFM, throughput is limited only by the ability to resolve individual tethered particles. In our instrument, image quality is limited by the camera, which was selected to provide exceptional value for financial and weight budgets: the image quality is sufficient for bead detection and the “binary” decision about the presence or absence of a bead (see below); the analog radio-frequency signal can be captured and digitized at video rates in real time; the cost of the camera is only $25 USD at the time of this writing; and the camera mass is only 9 g. Moreover, because each camera unit has a unique radio frequency output, further multiplexing of experiments would involve simply cloning MR.CFM with a different camera unit, and capturing video streams to the computer from parallel experiments within one centrifuge.
A wide field of view and high-efficiency surface chemistry enable the simultaneous visualization of hundreds of particles. High-throughput analysis is enabled by manipulating the raw image stream acquired by MR.CFM and computationally detecting and counting scores of particles (Fig. 3). The image processing used in this work minimizes user input and is implemented within a semi-automated ImageJ macro. With the use of this macro, beads occupying only a few pixels in the wide-field images from MR.CFM can be easily detected.
Collagen proteolysis under force
To determine whether tension affects stability of collagen’s triple helix, we determined how an applied force alters its rate of cleavage by trypsin. Single type III collagen molecules were tethered by a heavy microsphere to a surface, and force was applied from gravity (F = 0.067 pN ≈ 0) or by force from rotation of a centrifuge (Fig. 2 d). Tether-rupture kinetics were measured in the presence and absence of trypsin. Movie S1 provides an example experimental data set with collagen strained by 9 pN of force, in the presence of trypsin.
From measurements on thousands of distinct single-molecule tethers, our data show a significantly enhanced rate of collagen cleavage by trypsin in the presence of applied force (Figs. 4 and S5). For each experimental condition, the fraction of beads remaining as a function of time, f(t), was well described by a single-exponential decay process. Thus, the data were fit to obtain first-order effective rate constants:
| (1) |
Bead loss in the absence of trypsin was ascribed to nonspecific detachment, such that keff = kns. In the presence of trypsin, two competing processes contribute to bead loss, nonspecific detachment and trypsin-dependent cleavage; thus, keff = kns + kTr. Both kns and kTr depend on force, giving
| (2a) |
| (2b) |
where the index i = 0 for the no-force experiments and i = F for experiments under 9 pN of force. fo represents the fraction of beads that do not detach and can vary between each experimental data set. Equation 2b arises directly from the following first-order decay model:
| (3) |
Rate constants resulting from this analysis are reported in Table 2.
Our data show a substantial increase in the trypsin-dependent cleavage rate as the force increased from 0 to 9 pN: from kTr,0 = 0.009 ± 0.013 min−1 to kTr,F = 0.222 ± 0.018 min−1. These values were determined by fitting the mean fraction of beads remaining as a function of time (determined from multiple experimental replicates), weighted by the standard error of the mean fraction at each point. Alternative approaches to the data analysis similarly found a significant increase in the rate of collagen cleavage by trypsin (Tables S2 and S3), with rates enhanced 10- to 20-fold at 9 pN of force compared to the no-load condition. This result suggests that as collagen is stretched, the triple helix becomes more susceptible to cleavage by trypsin.
To confirm that the effect of force is on cleavage of collagen rather than of other proteins in the system used as linkers, control experiments were performed. Both streptavidin and BSA, used in the linking chemistry to connect collagen to the surface, have multiple trypsin recognition sites. Thus, the force-enhanced cleavage rate could arise from force-induced destabilization of streptavidin and/or BSA rather than of collagen. To investigate this possibility, we repeated the above experiments but used DNA as a tether in place of collagen. DNA is not a substrate for trypsin; therefore, any trypsin-dependent cleavage observed would arise from cleavage of the linking proteins. These experiments showed no force enhancement of the trypsin-dependent cleavage rate, kTr (Fig. S4). This result implies that the linking streptavidin and BSA are not destabilized by force exerted through the molecular tether. Thus, these findings indicate that the enhancement of proteolysis is due to a destabilization of collagen’s triple helix when stretched, which increases the accessibility of otherwise sterically hindered chains to the protease active site.
The destabilization of collagen’s triple helix by force can be compared with previous findings (Table 1). It is possible that shear-induced helix rupture (10) contributes to our force-enhanced bead detachment in the absence of trypsin. However, the loss of tethered beads from the surface, even in the absence of enzyme, does not occur with different surface chemistry (Fig. S6) and so is more likely attributed to mechanically weak linking interactions, perhaps between biotinylated BSA and silane (18).
Force-enhanced cleavage is consistent with conclusions from single-molecule collagen studies that used the collagenase matrix-metalloprotease-1 (MMP1) (9, 15), but contradictory to those from studies that used bacterial collagenase (14). Because these enzymes target different sequences of collagen, it is possible that collagen exhibits a sequence-dependent response to applied load. Interestingly, of the 81 possible trypsin sites per type III collagen chain, previous analysis of collagen fragments produced by trypsin proteolysis identified one primary cleavage site (25) located in a “loose” region of the triple helix that is shared by the unique recognition sequence of MMPs (26). The force enhancement of collagen proteolysis by trypsin, as by MMP1, may be a result of both enzymes probing the same region of the collagen triple helix. Indeed, our finding of a non-zero cleavage rate at zero force is consistent with the expectation that type III collagen is weakly susceptible to trypsin digestion in this MMP region in solution at room temperature (25). In contrast to MMP1, trypsin has not evolved to interact with triple-helical collagen and thus may be considered a more passive probe of externally induced changes in collagen’s structure.
Just as distinct sequences of triple helix can possess different thermal stabilities and helical pitch (5), it is possible that collagen has a molecular structure whose response to applied force is modulated by its local sequence. Future experiments could examine this question by using different collagen substrates, such as type I collagen that lacks a trypsin site in the MMP target region, or by using different proteases to target distinct regions of collagen’s sequence, thereby mapping the sequence-dependent mechanical stability within the native, full-length protein. How distinct mechanical signatures along the length of each collagen interact with and modulate biochemical and cell biological interactions is an exciting question to pose for future studies. More generally, the real-time, highly multiplexed approach of MR.CFM enables the discovery and characterization of transiently accessible and mechanically cryptic sites within native states of proteins (27), protein-DNA structures, and protein assemblies.
Data availability
The data sets generated and analyzed during the current study are available from the corresponding author on request.
Author Contributions
M.W.H.K. and N.R.F. designed the research. M.W.H.K. built MR.CFM and performed all experiments. M.W.H.K. and N.R.F. analyzed the results and wrote the manuscript.
Acknowledgments
We are grateful to Rachel Altman for insightful discussions regarding data analysis, Aaron Lyons for AFM imaging and analysis, Wesley Wong and Ken Halvorsen for helpful discussions, and David Lee for the use of the bright-field microscope. We thank John Bechhoefer, Edgar Young, and members of the Forde laboratory for critical readings of the manuscript.
This research was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC). M.W.H.K. acknowledges support from Robert Russell Family/First Nations Graduate Awards.
Editor: Alexander Dunn.
Footnotes
Supporting Materials and Methods, six figures, four tables, one movie, and two data files are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)35085-3.
Supporting Citations
Reference (28) appears in the Supporting Material.
Supporting Material
Collagen cleavage by trypsin under force. Video presents the filtered image stream used for data analysis and represents 20 min of data.
Includes technical drawings of MR.CFM lens adaptor, MR.CFM metal insert, and MR.CFM plastic base.
References
- 1.Bruckner P., Prockop D.J. Proteolytic enzymes as probes for the triple-helical conformation of procollagen. Anal. Biochem. 1981;110:360–368. doi: 10.1016/0003-2697(81)90204-9. [DOI] [PubMed] [Google Scholar]
- 2.Polach K.J., Widom J. Restriction enzymes as probes of nucleosome stability and dynamics. Methods Enzymol. 1999;304:278–298. doi: 10.1016/s0076-6879(99)04017-3. [DOI] [PubMed] [Google Scholar]
- 3.Park C., Marqusee S. Probing the high energy states in proteins by proteolysis. J. Mol. Biol. 2004;343:1467–1476. doi: 10.1016/j.jmb.2004.08.085. [DOI] [PubMed] [Google Scholar]
- 4.Mortimer G.M., Minchin R.F. Cryptic epitopes and functional diversity in extracellular proteins. Int. J. Biochem. Cell Biol. 2016;81:112–120. doi: 10.1016/j.biocel.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 5.Shoulders M.D., Raines R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang S.W., Buehler M.J. Molecular biomechanics of collagen molecules. Mater. Today. 2014;17:70–76. [Google Scholar]
- 7.Tonge T.K., Ruberti J.W., Nguyen T.D. Micromechanical modeling study of mechanical inhibition of enzymatic degradation of collagen tissues. Biophys. J. 2015;109:2689–2700. doi: 10.1016/j.bpj.2015.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung L., Dinakarpandian D., Nagase H. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 2004;23:3020–3030. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adhikari A.S., Chai J., Dunn A.R. Mechanical load induces a 100-fold increase in the rate of collagen proteolysis by MMP-1. J. Am. Chem. Soc. 2011;133:1686–1689. doi: 10.1021/ja109972p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zitnay J.L., Li Y., Weiss J.A. Molecular level detection and localization of mechanical damage in collagen enabled by collagen hybridizing peptides. Nat. Commun. 2017;8:14913. doi: 10.1038/ncomms14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varma S., Botlani M., Schieber J.D. Effect of intrinsic and extrinsic factors on the simulated D-band length of type I collagen. Proteins. 2015;83:1800–1812. doi: 10.1002/prot.24864. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y.L., Luo Z.P., An K.N. Direct quantification of the flexibility of type I collagen monomer. Biochem. Biophys. Res. Commun. 2002;295:382–386. doi: 10.1016/s0006-291x(02)00685-x. [DOI] [PubMed] [Google Scholar]
- 13.Wieczorek A., Rezaei N., Forde N.R. Development and characterization of a eukaryotic expression system for human type II procollagen. BMC Biotechnol. 2015;15:112. doi: 10.1186/s12896-015-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camp R.J., Liles M., Ruberti J.W. Molecular mechanochemistry: low force switch slows enzymatic cleavage of human type I collagen monomer. J. Am. Chem. Soc. 2011;133:4073–4078. doi: 10.1021/ja110098b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adhikari A.S., Glassey E., Dunn A.R. Conformational dynamics accompanying the proteolytic degradation of trimeric collagen I by collagenases. J. Am. Chem. Soc. 2012;134:13259–13265. doi: 10.1021/ja212170b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckhard U., Schönauer E., Brandstetter H. Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nat. Struct. Mol. Biol. 2011;18:1109–1114. doi: 10.1038/nsmb.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sbalzarini I.F., Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Hua B., Han K.Y., Ha T. An improved surface passivation method for single-molecule studies. Nat. Methods. 2014;11:1233–1236. doi: 10.1038/nmeth.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shayegan M., Rezaei N., Forde N.R. Probing multiscale mechanics of collagen with optical tweezers. In: Dholakia K., Spalding G.C., editors. Proceedings of the SPIE. SPIE; 2013. pp. 88101–88110. [Google Scholar]
- 20.Gasteiger E., Hoogland C., Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 21.Halvorsen K., Wong W.P. Massively parallel single-molecule manipulation using centrifugal force. Biophys. J. 2010;98:L53–L55. doi: 10.1016/j.bpj.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoang T., Patel D.S., Halvorsen K. A wireless centrifuge force microscope (CFM) enables multiplexed single-molecule experiments in a commercial centrifuge. Rev. Sci. Instrum. 2016;87:083705. doi: 10.1063/1.4961477. [DOI] [PubMed] [Google Scholar]
- 24.Yang D., Ward A., Wong W.P. Multiplexed single-molecule force spectroscopy using a centrifuge. Nat. Commun. 2016;7:11026. doi: 10.1038/ncomms11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller E.J., Finch J.E., Jr., Robertson P.B. Specific cleavage of the native type III collagen molecule with trypsin. Similarity of the cleavage products to collagenase-produced fragments and primary structure at the cleavage site. Arch. Biochem. Biophys. 1976;173:631–637. doi: 10.1016/0003-9861(76)90300-3. [DOI] [PubMed] [Google Scholar]
- 26.Fields G.B. A model for interstitial collagen catabolism by mammalian collagenases. J. Theor. Biol. 1991;153:585–602. doi: 10.1016/s0022-5193(05)80157-2. [DOI] [PubMed] [Google Scholar]
- 27.Gordon W.R., Zimmerman B., Blacklow S.C. Mechanical allostery: evidence for a force requirement in the proteolytic activation of Notch. Dev. Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shayegan M., Altindal T., Forde N.R. Intact telopeptides enhance interactions between collagens. Biophys. J. 2016;111:2404–2416. doi: 10.1016/j.bpj.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Collagen cleavage by trypsin under force. Video presents the filtered image stream used for data analysis and represents 20 min of data.
Includes technical drawings of MR.CFM lens adaptor, MR.CFM metal insert, and MR.CFM plastic base.
Data Availability Statement
The data sets generated and analyzed during the current study are available from the corresponding author on request.