Abstract
Intramembrane aspartyl proteases (IAPs) comprise one of four families of integral membrane proteases that hydrolyze substrates within the hydrophobic lipid bilayer. IAPs include signal peptide peptidase, which processes remnant signal peptides from nascent polypeptides in the endoplasmic reticulum, and presenilin, the catalytic component of the γ-secretase complex that processes Notch and amyloid precursor protein. Despite their broad biomedical reach, basic structure-function relationships of IAPs remain active areas of research. Characterization of membrane-bound proteins is notoriously challenging due to their inherently hydrophobic character. For IAPs, oligomerization state in solution is one outstanding question, with previous proposals for monomer, dimer, tetramer, and octamer. Here we used small angle neutron scattering (SANS) to characterize n-dodecyl-β-D-maltopyranoside (DDM) detergent solutions containing and absent a microbial IAP ortholog. A unique feature of SANS is the ability to modulate the solvent composition to mask all but the enzyme of interest. The signal from the IAP was enhanced by deuteration and, uniquely, scattering from DDM and buffers were matched by the use of both tail-deuterated DDM and D2O. The radius of gyration calculated for IAP and the corresponding ab initio consensus model are consistent with a monomer. The model is slightly smaller than the crystallographic IAP monomer, suggesting a more compact protein in solution compared with the crystal lattice. Our study provides direct insight into the oligomeric state of purified IAP in surfactant solution, and demonstrates the utility of fully contrast-matching the detergent in SANS to characterize other intramembrane proteases and their membrane-bound substrates.
Introduction
Intramembrane proteases (IPs) cleave membrane-embedded substrates within the confines of the lipid bilayer. Products of IPs are peptides and proteins involved in various biochemical processes such as cell metabolism, differentiation, development, immune response, and surveillance (1), and IPs are drug targets for a number of diseases (2). IPs are categorized by their nucleophile: serine, cysteine, aspartate, or their metal ion requirement (2, 3, 4, 5). Intramembrane aspartyl proteases include presenilin and signal peptide peptidase (SPP). Presenilin is the catalytic component of γ-secretase responsible for generating amyloid-β (Aβ) peptides implicated in Alzheimer’s disease from amyloid precursor protein (3, 5), and SPP cleaves remnant leader peptides in the endoplasmic reticulum membrane after cleavage by the soluble signal peptidase (6). In humans, SPP substrates include leader peptides from proteins involved in immune system, inflammatory response, and hepatitis C viral maturation (7, 8, 9). SPP and presenilin share key sequence and catalytic similarity, including signature catalytic motifs (YD and GxGD where x is any amino acid) and inhibition (10, 11, 12, 13); similarities extend to microbial orthologs (12, 13, 14).
Despite their broad biomedical reach, the structure of active intramembrane aspartyl proteases (IAPs) including the number of subunits in the functional enzyme, has remained ambiguous. Membrane proteins require the presence of a mild detergent or other amphiphilic system to solubilize and stabilize a given membrane protein in an active conformation, which hinders structural characterization using standard analytical techniques used for soluble proteins such as size exclusion chromatography or small angle x-ray scattering (SAXS). In the case of size exclusion chromatography, even coupled with multiangle light scattering, molecular mass determination is only possible when the protein-detergent complex is well separated from empty micelles (15), and in SAXS, the signal from the membrane protein cannot be isolated from that of the solubilizing agent (16). For IAPs, stoichiometries of one (17) through eight (18) subunits have been proposed. Before recent cryo-electron microscopy studies of γ-secretase that indicate a monomer (17), homodimer (19, 20, 21, 22, 23) was the predominant proposal. Fluorescence lifetime imaging microscopy was used to demonstrate that both SPP (21, 22) and presenilins (19, 20) are dimers. The lattice arrangement of the 3.8 Å resolution crystal structure of the microbial Methanoculleus marisnigri IAP (WP_011844759.1, MmIAP) ortholog suggests either a dimer or tetramer (24).
To date, no IP has been structurally characterized using small angle neutron scattering (SANS), a method well suited to study the solution properties of macromolecules and complex multicomponent assemblies like membrane proteins (25). For membrane protein-detergent complexes, the different scattering length densities of each component can be exploited so that only the membrane protein is visualized during the scattering experiment. Typically, the scattering length densities of the detergent and buffer/solvent are matched by judiciously adjusting the ratio of H2O and D2O in the solvent (26), and the signal from the membrane protein can be further enhanced by deuteration during cell growth (25).
Here we report the solution structure of deuterated MmIAP in n-dodecyl-β-d-maltopyranoside (DDM) determined by SANS. After deuterating MmIAP, the detergent was fully contrast-matched using a specific ratio of hydrogenated and tail-deuterated detergent, and the scattering profile for MmIAP was recorded. The radius of gyration (Rg) calculated for MmIAP from SANS is smaller than the Rg calculated from the crystallographic MmIAP monomer, suggesting a more compact protein in DDM-containing solution. Our SANS study provides, to our knowledge, new insight into the solution oligomeric state of MmIAP in detergent solution, and bolsters the utility of SANS to characterize other IPs and their membrane-bound substrates.
Materials and Methods
Expression and purification of protonated and deuterated MmIAP
The plasmid containing the MmIAP gene with a C-terminal hexahistidine tag was transformed into Escherichia coli Rosetta 2 cells (Novagen, Madison, WI). Cells were grown, membranes isolated, and protein purified as described previously for enzyme activity assays (14). Modest modifications were made to scale purification to the higher protein requirement for SANS experiments. Membrane (∼2 g) was solubilized in 160 mL of 50 mM Hepes (pH 7.5), 500 mM NaCl, 20 mM imidazole, and 1% (w/v) DDM (Anatrace, Maumee, OH) by gentle rocking for 1 h at 4°C. Unsolubilized material was removed via ultracentrifugation at 162,000 × g for 45 min. The supernatant containing solubilized membranes was loaded onto a 1 mL HisTrap FF Crude nickel affinity chromatography column (GE Healthcare, Chicago, IL) pre-equilibrated with Buffer A [50 mM Hepes (pH 7.5), 500 mM NaCl, 20 mM imidazole, 0.1% DDM]. Before elution of MmIAP, weakly bound impurities were removed with 5% Buffer B [50 mM Hepes (pH 7.5), 500 mM NaCl, 500 mM imidazole, 0.1% DDM]. Elution of purified MmIAP was accomplished using a linear gradient by mixing Buffer A and 5–60% Buffer B. Elution fractions containing MmIAP were pooled and further purified on a HiPrep 16/60 Sephacryl-S300 (GE Healthcare) using gel filtration buffer [20 mM Hepes (pH 7.5), 250 mM NaCl, 0.05% DDM]. Purity of protein was assessed by denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis (12% polyacrylamide) stained with Coomassie and concentration was measured using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA) (ε = 33,920 M−1 cm−1). Before SANS measurements, pure (protonated) MmIAP was buffer-exchanged using a 500 μL Amicon Ultra MWCO 50K concentrator (Millipore, Burlington, MA) into gel filtration buffer containing 22% D2O (Acros Organic, Geel, Belgium).
Fed-batch cultivation, expression of deuterated MmIAP (d-MmIAP), and cell lysis was carried out in the Bio-Deuteration Laboratory at Oak Ridge National Laboratory (ORNL, Oak Ridge, TN). Before d-MmIAP production, E. coli Rosetta 2 cells were first adapted to D2O by transferring an individual colony of transformed cells from a Luria Broth agar plate to 0.5% (w/v) glycerol minimal medium in H2O, and then subculturing into the same medium with an increasing D2O content (25, 50, 75, and 90%), 100 μg/mL carbenicillin, and 17 μg/mL chloramphenicol. Once the cells were growing in 90% D2O medium, a 400 mL preculture was used to inoculate 3.6 L of fresh 90% D2O medium in a benchtop BioFlo 310 bioreactor system (Eppendorf, Hauppauge, NY) equipped with a 5.5 L working volume vessel. At the outset of the experiment, the temperature was maintained at 30°C, aeration was maintained throughout at 4 L/min, and agitation was varied from 200 to 800 Rpm to maintain dissolved oxygen above a set point of 30% saturation. A solution of 10% (w/v) NaOH in 90% D2O was added on demand to maintain a pD >7.3. When the dissolved oxygen spike occurred upon depletion of the 0.5% (w/v) glycerol, a feeding of a solution consisting of 10% (w/v) glycerol and 0.2% MgSO4 in 90% D2O was initiated. After ∼7 h of feeding, the temperature set point was reduced to 18°C and d-MmIAP expression was induced by adding 1 mM isopropyl-β-D-1-thiogalactopyranoside. Upon harvesting via centrifugation at 6000 × g for 45 min, cell paste containing d-MmIAP (∼145 g wet cell weight) was suspended (0.1 g/mL) in 50 mM Hepes (pH 7.5) and 200 mM NaCl containing EDTA-Free SIGMAFAST Protease Inhibitor Cocktail Tablets (Sigma Aldrich, St. Louis, MO), and lysed at 15,000 psi via three passages through an EmulsiFlex-C3 homogenizer that was fitted with a chilled heat exchanger (Avestin, Ottawa, ON) and stationed in a 6°C cold room. After lysis, cellular debris was removed by centrifugation at 5000 × g for 15 min for four times. The supernatant (∼1.8 L) was then subjected to ultracentrifugation at 162,000 × g for 30 min. The pelleted membrane fraction was washed by resuspension in a Dounce homogenizer and the membrane fraction was again isolated by ultracentrifugation as above. d-MmIAP-containing membranes (∼22 g) were solubilized by gentle rocking for 0.5 h at 4°C in 200 mL of a solution containing 50 mM Hepes (pH 7.5), 500 mM NaCl, 20 mM imidazole, and 4% (w/v) DDM. The membrane resuspension was subjected to ultracentrifugation as above to remove insoluble material.
The supernatant containing the solubilized membrane was purified using the protocol for MmIAP described above except for the following additional steps to maximize yield of purified enzyme. Namely, the flow through fractions from the first nickel affinity chromatography run were diluted in Buffer A lacking detergent, to a final concentration of 2% DDM, and purified on the column again. A third round of nickel affinity chromatography was then performed on the flow through from the second column, which was diluted with Buffer A lacking detergent to 1% DDM. Elution fractions from the three nickel affinity purification runs were pooled, concentrated to two ∼900 μL aliquots using a 15 mL Amicon Ultra MWCO 50K concentrator (Millipore), and each loaded onto a HiPrep 16/60 Sephacryl-S300 (GE Healthcare) pre-equilibrated as for MmIAP above. Fractions containing purified d-MmIAP as judged by sodium dodecyl sulfate polyacrylamide gel electrophoresis were pooled and a third Sephacryl-S300 column was used to further polish the sample. In a final step, purified d-MmIAP was loaded onto a Superose-12 10/300 GL (GE Healthcare) column equilibrated with 20 mM Hepes (pH 7.5), 250 mM NaCl, 48.5% D2O, and 0.05% total DDM, of which 44% (w/v) is tail-deuterated d25-DDM (Anatrace) (Fig. S1). The final yield was ∼1 mg of d-MmIAP, all of which was used in the SANS experiment.
SANS data collection
SANS data were collected at the Bio-SANS beam line CG-3 of the High-Flux Isotope Reactor at ORNL using a single instrument configuration with 7-meter sample-to-detector distance. Data were collected at 12°C using 1 mm quartz cells and neutron wavelength of 6 Å ± 14%. The range of momentum transfer Q used was 0.007 < Q < 0.7 Å−1 (Q = 4π × sin(θ)/λ, where 2θ is the scattering angle and λ is the neutron wavelength). Additional descriptions of the instrument and setup have been previously published (27, 28, 29). The recorded scattering data were circularly averaged, and reduced to one-dimensional scattering profiles using MantidPlot software (30). Calibration of the SANS data to an absolute scale was performed by measuring a porous silica standard with known intensity at zero angle (extrapolated from a Debye-Bueche plot). Blank buffers containing the same percentage D2O as the samples were similarly measured and subtracted from the sample scattering for background correction using a toolkit developed by Dr. Ken Littrell (ORNL) for IgorPro. Subsequent data analysis and modeling of scattering profiles were facilitated with the ATSAS 2.6.1 program suite (European Molecular Biology Laboratory, Hamburg) (31).
SANS data analysis and modeling
An initial estimation of the Rg and forward scattering intensity (I0) was performed using PRIMUS (32). Core-shell ellipsoid shape models were fit to the scattering data using SasView v4.1 (33). Comparisons of SANS data for d-MmIAP to structures of MmIAP and presenilin in the PDB were conducted with CRYSON (34). For the simulation conditions, a deuteration fraction of 0.75 was used for the protein chain with a D2O fraction in the solvent of 0.49 to mimic the experimental contrast conditions.
ATSAS software tools were used for further modeling and interpretation of the structural SANS information for d-MmIAP in solution with contrast-matched detergent. The first step of this process employed GNOM (35) to generate a pair-distance distribution function P(r), which described the relative interatomic distances within the scattering particle. Scattering data over the range 0.0106 < Q < 0.585 Å−1 were used for the real space transformation and subsequent modeling, and provided a P(r) curve with a single peak and Dmax of 46 Å. The Rg from the real space transformation was 16.7 ± 0.2 Å with an I0 of 0.610 ± 0.001 cm−1. The GNOM output was used for ab initio molecular shape generation with DAMMIF and DAMMIN (36, 37). The P(r) data were used as an input for fast DAMMIF modeling to create 17 initial dummy atom models (DAMs). These initial DAMs were aligned and averaged using DAMAVER (38), and one outlier from the 17 was discarded based on normalized spatial discrepancy (NSD) values, as its NSD value exceeded two standard deviations from the cluster mean (cluster NSD: 0.708 ± 0.018; outlier NSD: 0.749). After averaging in DAMAVER, the “damstart” (fixed-core) model was used for refinement with DAMMIN, yielding a single refined SANS envelope. Superimpositions of the SANS envelope with crystal structures were performed using SUPCOMB (39), which minimizes NSD to find the best alignment of the two models.
Results
SANS analysis of d-MmIAP
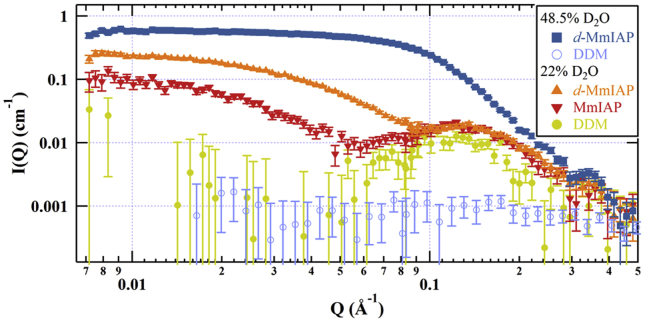
SAXS measurements showed, as expected, strong scattering signal from DDM micelles that was similar with and without MmIAP, and initial attempts of SANS experiments using nondeuterated MmIAP, DDM, and D2O/H2O were unsuccessful in isolating a signal for MmIAP unperturbed by scattering contributions from the surfactant (Fig. S2). Therefore, d-MmIAP was expressed and purified (Fig. S1) immediately before data collection, using established methods yielding active enzyme (14). Based on other studies (40), the average deuteration level under growth conditions is 70–75%. Scattering from the d-MmIAP protein-detergent complex produced a stronger signal overall as a result of the increased protein contrast relative to the solvent, but contributions from the detergent were still present in the net scattering profile. These results are observed in a comparison of the MmIAP-DDM complex versus d-MmIAP-DDM in the same DDM contrast-matched conditions (Fig. 1).
Figure 1.
SANS contrast match point measurements for DDM micelles (yellow, ●), MmIAP with DDM (red, ▼), d-MmIAP with DDM (orange, ▲) in 22% D2O, and mixed micelles (blue, ○) or d-MmIAP with DDM/d25-DDM mixed micelles (blue, ▪) in 48.5% D2O. Data for contrast-matched DDM micelles in 48.5% D2O was reproduced from (41). To see this figure in color, go online.
To achieve true extinction of any scattering contribution from DDM, a more refined approach was required. The individual contrast match points (CMPs) of hydrophobic DDM alkyl tails and hydrophilic maltoside headgroups are 2 and 48.5% D2O, respectively, which are very far from the overall CMP of 22% D2O. This, together with the similar size of both moieties and their distinctive location in a micelle core and shell produces significant residual scattering, even at 22% D2O (Fig. 1). This problem can be resolved by raising the CMP of the DDM micelle core to 48.5% D2O to match the shell by precisely blending 44% (w/v) tail-deuterated DDM (d25-DDM) with regular DDM (41). Under these complete matching conditions, scattering features from DDM micelles were rendered negligible (Fig. 1), which is readily apparent by the absence of a secondary maximum in the SANS profile.
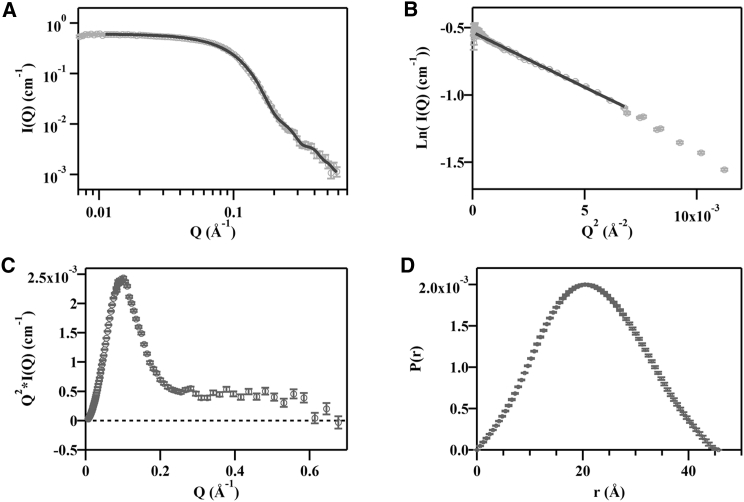
The combination of deuterated protein and completely contrast-matched mixed micelles yielded an interpretable SANS profile for the enzyme without interference from buffer and detergent components (Figs. 1 and 2 A). Guinier analysis was performed on the low-Q scattering data defined by an upper limit of Q × Rg < 1.3, and provided estimates of I0 and Rg (Fig. 2 B). The measured Rg (16.1 ± 0.5 Å), and a Kratky plot illustrating the folded nature of the scattering object (Fig. 2 C), suggests that MmIAP is most likely monomeric with a compact, globular shape in detergent solution (42). The forward scattering intensity determined from the Guinier fit was 0.60 ± 0.01 cm−1. An indirect Fourier transform of the scattering data provided a plot describing the pair-distance distribution P(r) of intramolecular distances within the particle, constrained by a maximum particle dimension of 46 Å (Fig. 2 D). Molecular weight was estimated from the DAMMIN model and by the method described by Rambo and Tainer (43) (see Supporting Material for details). A summary of the physical parameters extracted from the SANS data in comparison with similar values obtained from the final SANS envelope and the PDB model (4HYC, chain A) is presented in Table 1. The combination of these results from SANS suggest a monomeric MmIAP size and shape without the formation of higher order oligomers in micellar solution.
Figure 2.
SANS data obtained for d-MmIAP with contrast-matched DDM/d25-DDM mixed micelles (48.5% D2O). (A) A plot of the scattered intensity versus Q, a function of the scattering angle, with GNOM fit shown in solid line. (B) A Guinier plot of the low angle scattering data with a linear fit in the Guinier region shown as a solid line. (C) A Kratky plot of the same scattering data demonstrating the compact, folded shape of the particle. (D) A pair-distance distribution function P(r) obtained from GNOM representing the distribution of real space distances between scattering centers within the particle.
Table 1.
Summary of Physical Parameters from the SANS Data, Reconstructed DAM, and the Related PDB: 4HYC:A
| Radius of Gyration (Å) | Dmax (Å) | Volume (Å3) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Guinier | GNOM | DAM | PDB | GNOM | DAM | PDB | SANS | DAM | PDB |
| 16.1 ± 0.5 | 16.7 ± 0.2 | 16.7 | 19.4 | 46.0 | 47.7 | 72.9 | 21,030 | 20,590 | 31,258 |
Comparison with crystal structure and ab initio modeling
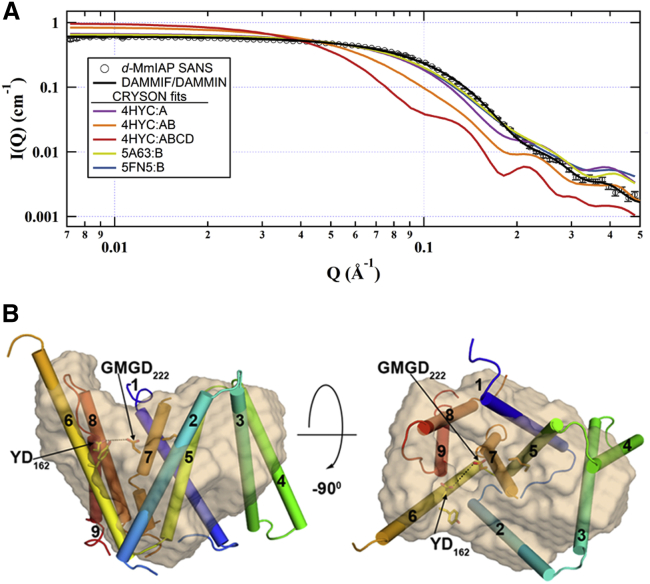
SANS profiles were calculated from available structures for pertinent enzymes (chain A from MmIAP PDB: 4HYC, 4HYD, 4HYG, and 4Y6K, and chain B, presenilin from γ-secretase PDB: 5A63, 5FN2, 5FN3, 5FN4, and 5FN5). The four MmIAP crystal structures are ∼3.3–3.9 Å resolution, represent three different space groups, different bound states (apo and inhibited), and are similar to each other with root-mean square deviation (RMSD) of ∼0.5–0.8 Å. The five presenilin structures solved by cryo-electron microscopy are 4.0–4.3 Å resolution, and represent four apo states, as well as one that is bound to an inhibitor. Presenilin shares just 15% identity with MmIAP yet pairwise RMSDs for the two different structures are ∼2.5–3 Å. CRYSON fits to the experimental data from representative members of each PDB group are shown in Fig. 3 A. The simulated scattering profiles for structural monomers are in relative agreement with the measured scattering profile of d-MmIAP in contrast-matched, mixed d25-DDM/DDM micelles (Fig. 3). Each structure appears somewhat larger than that measured in solution, denoted by the decrease in scattered intensity from the plateau at lower values of Q for the simulated SANS data compared with the measured data, but are much closer to the data than a hypothetical d-MmIAP dimer and tetramer (Fig. 3 A). The average Rg from the structure (19.4 Å, Table 1) is also ∼3.4 Å larger than the Rg determined from a Guinier fit to the SANS data (16.1 ± 0.5 Å).
Figure 3.
CRYSON results and comparison with SANS data. (A) Simulated SANS profiles from CRYSON for representative PDB entries related to IAPs. Scattering from a possible MmIAP dimer and tetramer were simulated using chains A and B from 4HYC, or chains A–D from 4HYC, respectively, demonstrating that higher ordered oligomers are not consistent with the observed scattering. (B) The ab initio model was overlaid with chain A from 4HYC to provide a three-dimensional structural comparison. TM helices numbered from 1 (N-terminus (blue)) to 9 (C-terminus (red)), and catalytic motif YD. GMGD is represented as sticks. To see this figure in color, go online.
Ab initio modeling from d-MmIAP SANS data with contrast-matched detergent recapitulates the overall agreement with the available structures. A representative scattering profile from the remaining DAMMIF models demonstrates strong agreement with the experimental data (Fig. 3 A) and the SANS profile of the DAMMIN-refined envelope was indistinguishable from this representative trace. The SANS envelope shown here has a protrusion that is consistent with the predicted position of the long helix 6, which contains the C-terminal YD motif, and a well leading to the general vicinity of the catalytic aspartates (Fig. 3 B). However, we note that not all 16 DAMs show exactly these features in the same location; thus, the fit shown here agrees with the SANS data, but this envelope is not the only possible model at this level of detail. The poorest fit is for helix 4, half of which appears to protrude beyond the SANS envelope in accordance with the larger Rg and Dmax values (Table 1) and calculated scattering intensity at low-Q values.
Discussion
Despite the increasing number of available membrane protein structures in the protein data bank determined by x-ray crystallography, NMR spectroscopy, and cryo-electron microscopy, the percentage remains very low compared with the total membrane-bound proteome, and determining the oligomerization state of such proteins in solution remains a major challenge (44, 45, 46). Here we used SANS to determine the molecular envelope and oligomerization state of an IAP in solution. Our strategy had three major components, 1) deuterating MmIAP during cell growth to increase the SANS signal from the enzyme, 2) utilizing 44% d25-DDM to match the CMP of the hydrophobic tail core with that of the headgroup of the DDM micelle, and 3) contrast matching the overall DDM micelle with 48.5% D2O to leave only scattering from d-MmIAP.
The process of contrast matching to detect only the signal from the membrane protein is inherently challenging (47); micelle-micelle interaction or protein-protein interactions can corrupt low-Q data, and the residual signal from incompletely matched lipid head and tail components limit data interpretation to gross structural changes under defined experimental conditions. Typically, SANS studies of membrane proteins employ the average CMP for the solubilizing agent. For example, Bu et al. (48) overall contrast-matched the small unilamellar vesicles to detect a change in the oligomeric state of SecA upon nucleotide binding, and Zimmer et al. (49) used the average contrast match for DDM of 22% D2O to detect differences between truncated and full-length potassium channel KcsA solubilized in DDM under different pH conditions. With our improved understanding of the detergent contributions to neutron scattering (41), reinspection of numerous SANS studies that utilize a similar strategy (49, 50, 51, 52, 53, 54) suggests that the scattering profile exhibits contributions due to incompletely matched surfactant or lipid components. Thus, our recent efforts have been aimed at improving the contrast matching protocol by matching the hydrophobic tail of the detergent to its headgroup before contrast matching the overall micelle with D2O. One prior study used an analogous strategy with sodium dodecyl sulfate (55), but to our knowledge, our study is the first such application to study a membrane enzyme solubilized in DDM, a milder detergent with a larger headgroup (45). The theoretical basis for the approach with DDM was recently reported (41), and resulted in a strong interpretable signal free of contribution from the detergent.
The d-MmIAP ab initio model in solution is consistent with an approximately spherical monomeric protein, not a dimer or higher ordered species suggested in earlier biochemical experiments for SPP and other human IAP family members. Interestingly, the experimentally determined Rg (16.1 ± 0.5 Å) from SANS is somewhat smaller than the calculated Rg (19.4 Å) from a monomer chain of crystalline MmIAP, indicating a more compact structure is present in solution. This finding agrees with the observation that crystallized MmIAP was trapped in an inactive conformation, with the two catalytic aspartates too far apart for catalysis. Detergent identity and concentration are well known to affect crystallization properties (56, 57). In the case of the MmIAP crystal structure, perhaps the limited proteolytic digestion of the enzyme or mutations introduced to enhance crystallizability (24), led to a less compact bundle of transmembrane helices. Alternatively, the dynamic detergent micelles present during crystallization might affect the lattice (58, 59, 60), as would be expected for an α-helical membrane protein with predominantly membrane-immersed helices (61). Our sample for SANS was prepared using the same methods as for our enzymatic study where it is active for nearly a week after purification (14), suggesting our SANS envelope reflects that of an active enzyme in detergent solution, but whether the protein remains a monomer in the presence of substrate is an open question for further study.
Conclusions
SANS analysis of d-MmIAP measured with fully contrast-matched detergent yields a molecular envelope consistent with a monomeric enzyme in solution. Comparison of the Rg value from SANS and previously reported crystal structure of an inactive IAP suggests the protein might form a more compact protein in solution, namely, tighter packing of the hydrophobic transmembrane helices. The finding that MmIAP is a monomer is consistent with the recent cryo-electron microscopy structure of presenilin in the context of γ-secretase, but differs from other experiments conducted with indirect biophysical or biochemical methods. The SANS approach and CMP strategy, used to study d-MmIAP in solution, should be applicable to study IAP structures in complex with substrates, other IP family proteins, and other well-behaved, detergent-solubilized membrane proteins.
Author Contributions
Conceptualization, Methodology, R.C.O., V.S.U., K.L.W., and R.L.L.; Investigation, S.-H.N., R.C.O., and K.L.W.; Writing – Original Draft, S.-H.N., R.C.O., V.S.U., and R.L.L.; Writing – Review and Editing, S.-H.N., R.C.O., K.L.W., V.S.U., and R.L.L.; Funding Acquisition and Supervision, R.L.L., and V.S.U.
Acknowledgments
This work was supported by a grant to R.L.L. from the National Science Foundation (NSF) (0845445). S.-H.N. acknowledges a travel grant from the College of Sciences, Georgia Tech to collect data at Oak Ridge National Laboratory. Neutron scattering studies at the CG-3 Bio-SANS instrument at the High-Flux Isotope Reactor and work in the Bio-Deuteration Laboratory of Oak Ridge National Laboratory were sponsored by the Office of Biological and Environmental Research and by the Scientific User Facilities Division, Office of Basic Energy Sciences, US Department of Energy (DOE). This work benefited from the use of the SasView application, originally developed under NSF award DMR-0520547. SasView also contains code developed with funding from the EU Horizon 2020 programme under the SINE2020 project grant 654000. This manuscript has been coauthored by UT-Battelle, LLC, under contract DE-AC05-00OR22725 with the DOE.
The US government retains and the publisher, by accepting the article for publication, acknowledges that the US government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for US government purposes. DOE will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Editor: James Cole.
Footnotes
Swe-Htet Naing and Ryan C. Oliver contributed equally to this work.
Supporting Materials and Methods and three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)35097-X.
Contributor Information
Volker S. Urban, Email: urbanvs@ornl.gov.
Raquel L. Lieberman, Email: raquel.lieberman@chemistry.gatech.edu.
Supporting Citations
Reference (62) appears in the Supporting Material.
Supporting Material
References
- 1.Brown M.S., Ye J., Goldstein J.L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Verhelst S.H.L. Intramembrane proteases as drug targets. FEBS J. 2017;284:1489–1502. doi: 10.1111/febs.13979. [DOI] [PubMed] [Google Scholar]
- 3.Urban S., Freeman M. Intramembrane proteolysis controls diverse signalling pathways throughout evolution. Curr. Opin. Genet. Dev. 2002;12:512–518. doi: 10.1016/s0959-437x(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 4.Manolaridis I., Kulkarni K., Barford D. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature. 2013;504:301–305. doi: 10.1038/nature12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erez E., Fass D., Bibi E. How intramembrane proteases bury hydrolytic reactions in the membrane. Nature. 2009;459:371–378. doi: 10.1038/nature08146. [DOI] [PubMed] [Google Scholar]
- 6.Weihofen A., Binns K., Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 7.Lemberg M.K., Bland F.A., Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. 2001;167:6441–6446. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- 8.Fluhrer R., Grammer G., Haass C. A gamma-secretase-like intramembrane cleavage of TNFalpha by the GxGD aspartyl protease SPPL2b. Nat. Cell Biol. 2006;8:894–896. doi: 10.1038/ncb1450. [DOI] [PubMed] [Google Scholar]
- 9.McLauchlan J., Lemberg M.K., Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weihofen A., Lemberg M.K., Martoglio B. Targeting presenilin-type aspartic protease signal peptide peptidase with gamma-secretase inhibitors. J. Biol. Chem. 2003;278:16528–16533. doi: 10.1074/jbc.M301372200. [DOI] [PubMed] [Google Scholar]
- 11.Sato T., Nyborg A.C., Wolfe M.S. Signal peptide peptidase: biochemical properties and modulation by nonsteroidal antiinflammatory drugs. Biochemistry. 2006;45:8649–8656. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- 12.Torres-Arancivia C., Ross C.M., Ubarretxena-Belandia I. Identification of an archaeal presenilin-like intramembrane protease. PLoS One. 2010;5:e13072. doi: 10.1371/journal.pone.0013072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang S., Wu S., Shi Y. Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH. Proc. Natl. Acad. Sci. USA. 2015;112:3344–3349. doi: 10.1073/pnas.1502150112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naing S.H., Vukoti K.M., Lieberman R.L. Catalytic properties of intramembrane aspartyl protease substrate hydrolysis evaluated using a FRET peptide cleavage assay. ACS Chem. Biol. 2015;10:2166–2174. doi: 10.1021/acschembio.5b00305. [DOI] [PubMed] [Google Scholar]
- 15.Gimpl K., Klement J., Keller S. Characterising protein/detergent complexes by triple-detection size-exclusion chromatography. Biol. Proced. Online. 2016;18:4. doi: 10.1186/s12575-015-0031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez J., Koutsioubas A. Memprot: a program to model the detergent corona around a membrane protein based on SEC-SAXS data. Acta Crystallogr. D Biol. Crystallogr. 2015;71:86–93. doi: 10.1107/S1399004714016678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai X.C., Yan C., Shi Y. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyashita H., Maruyama Y., Iwatsubo T. Three-dimensional structure of the signal peptide peptidase. J. Biol. Chem. 2011;286:26188–26197. doi: 10.1074/jbc.M111.260273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cervantes S., Saura C.A., Marfany G. Functional implications of the presenilin dimerization: reconstitution of gamma-secretase activity by assembly of a catalytic site at the dimer interface of two catalytically inactive presenilins. J. Biol. Chem. 2004;279:36519–36529. doi: 10.1074/jbc.M404832200. [DOI] [PubMed] [Google Scholar]
- 20.Herl L., Lleo A., Berezovska O. Detection of presenilin-1 homodimer formation in intact cells using fluorescent lifetime imaging microscopy. Biochem. Biophys. Res. Commun. 2006;340:668–674. doi: 10.1016/j.bbrc.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 21.Nyborg A.C., Herl L., Golde T.E. Signal peptide peptidase (SPP) dimer formation as assessed by fluorescence lifetime imaging microscopy (FLIM) in intact cells. Mol. Neurodegener. 2006;1:16. doi: 10.1186/1750-1326-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyborg A.C., Kornilova A.Y., Golde T.E. Signal peptide peptidase forms a homodimer that is labeled by an active site-directed gamma-secretase inhibitor. J. Biol. Chem. 2004;279:15153–15160. doi: 10.1074/jbc.M309305200. [DOI] [PubMed] [Google Scholar]
- 23.Schroeter E.H., Ilagan M.X., Kopan R. A presenilin dimer at the core of the gamma-secretase enzyme: insights from parallel analysis of Notch 1 and APP proteolysis. Proc. Natl. Acad. Sci. USA. 2003;100:13075–13080. doi: 10.1073/pnas.1735338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Dang S., Shi Y. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493:56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- 25.Breyton C., Gabel F., Ebel C. Small angle neutron scattering for the study of solubilised membrane proteins. Eur Phys J E Soft Matter. 2013;36:71. doi: 10.1140/epje/i2013-13071-6. [DOI] [PubMed] [Google Scholar]
- 26.Trewhella J. Neutrons reveal how nature uses structural themes and variation in biological regulation. Physica B. 2006;385–386:825–830. [Google Scholar]
- 27.Lynn G.W., Heller W., Myles D.A.A. Bio-SANS - A dedicated facility for neutron structural biology at oak ridge national laboratory. Physica B. 2006;385–386:880–882. [Google Scholar]
- 28.Heller W.T., Urban V.S., Myles D.A. The Bio-SANS instrument at the high flux isotope reactor of oak ridge ational laboratory. J. Appl. Cryst. 2014;47:1238–1246. [Google Scholar]
- 29.He L.L., Do C., Smith G.S. Corrections for the geometric distortion of the tube detectors on SANS instruments at ORNL. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrom. Detect. Assoc. Equip. 2015;775:63–70. [Google Scholar]
- 30.Arnold O., Bilheux J.C., Zikoysky J. Mantid-Data analysis and visualization package for neutron scattering and mu SR experiments. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrom. Detect. Assoc. Equip. 2014;764:156–166. [Google Scholar]
- 31.Petoukhov M.V., Franke D., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konarev P.V., Volkov V.V., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 33.Doucet, M., J. H. Cho, …, A. Washinton. 2017. SasView version 4.1. http://www.sasview.org/.
- 34.Svergun D.I., Richard S., Zaccai G. Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. USA. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 36.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volkov V.V., Svergun D.I. Uniqueness of ab initioshape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. [Google Scholar]
- 39.Kozin M.B., Svergun D.I. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 40.Leiting B., Marsilio F., O’Connell J.F. Predictable deuteration of recombinant proteins expressed in Escherichia coli. Anal. Biochem. 1998;265:351–355. doi: 10.1006/abio.1998.2904. [DOI] [PubMed] [Google Scholar]
- 41.Oliver R.C., Pingali S.V., Urban V.S. Designing mixed detergent micelles for uniform neutron contrast. J. Phys. Chem. Lett. 2017;8:5041–5046. doi: 10.1021/acs.jpclett.7b02149. [DOI] [PubMed] [Google Scholar]
- 42.Glatter O., Kratky O. Academic Press; Cambridge, MA: 1982. Small Angle X-ray Scattering. [Google Scholar]
- 43.Rambo R.P., Tainer J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker J.L., Newstead S. Membrane protein crystallisation: current trends and future perspectives. Adv. Exp. Med. Biol. 2016;922:61–72. doi: 10.1007/978-3-319-35072-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Privé G.G. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 46.le Maire M., Champeil P., Moller J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 47.Hunt J.F., McCrea P.D., Engelman D.M. Assessment of the aggregation state of integral membrane proteins in reconstituted phospholipid vesicles using small angle neutron scattering. J. Mol. Biol. 1997;273:1004–1019. doi: 10.1006/jmbi.1997.1330. [DOI] [PubMed] [Google Scholar]
- 48.Bu Z., Wang L., Kendall D.A. Nucleotide binding induces changes in the oligomeric state and conformation of Sec A in a lipid environment: a small-angle neutron-scattering study. J. Mol. Biol. 2003;332:23–30. doi: 10.1016/s0022-2836(03)00840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmer J., Doyle D.A., Grossmann J.G. Structural characterization and pH-induced conformational transition of full-length KcsA. Biophys. J. 2006;90:1752–1766. doi: 10.1529/biophysj.105.071175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cardoso M.B., Smolensky D., O’Neill H. Insight into the structure of light-harvesting complex II and its stabilization in detergent solution. J. Phys. Chem. B. 2009;113:16377–16383. doi: 10.1021/jp905050b. [DOI] [PubMed] [Google Scholar]
- 51.Johs A., Hammel M., Prassl R. Modular structure of solubilized human apolipoprotein B-100. Low resolution model revealed by small angle neutron scattering. J. Biol. Chem. 2006;281:19732–19739. doi: 10.1074/jbc.M601688200. [DOI] [PubMed] [Google Scholar]
- 52.Kumar-Singh S., Theuns J., Van Broeckhoven C. Mean age-of-onset of familial alzheimer disease caused by presenilin mutations correlates with both increased Abeta42 and decreased Abeta40. Hum. Mutat. 2006;27:686–695. doi: 10.1002/humu.20336. [DOI] [PubMed] [Google Scholar]
- 53.Nogales A., García C., González-Rodríguez J. Three-dimensional model of human platelet integrin alphaIIb beta3 in solution obtained by small angle neutron scattering. J. Biol. Chem. 2010;285:1023–1031. doi: 10.1074/jbc.M109.050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang K.H., Urban V.S., Blankenship R.E. SANS investigation of the photosynthetic machinery of Chloroflexus aurantiacus. Biophys. J. 2010;99:2398–2407. doi: 10.1016/j.bpj.2010.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clifton L.A., Johnson C.L., Lakey J.H. Low resolution structure and dynamics of a colicin-receptor complex determined by neutron scattering. J. Biol. Chem. 2012;287:337–346. doi: 10.1074/jbc.M111.302901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anandan A., Vrielink A. Detergents in membrane protein purification and crystallisation. Adv. Exp. Med. Biol. 2016;922:13–28. doi: 10.1007/978-3-319-35072-1_2. [DOI] [PubMed] [Google Scholar]
- 57.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 58.Grisshammer R. New approaches towards the understanding of integral membrane proteins: a structural perspective on G protein-coupled receptors. Protein Sci. 2017;26:1493–1504. doi: 10.1002/pro.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moraes I., Evans G., Stewart P.D.S. Membrane protein structure determination - the next generation. Biochim. Biophys. Acta. 2014;1838(1 Pt A):78–87. doi: 10.1016/j.bbamem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiener M.C. A pedestrian guide to membrane protein crystallization. Methods. 2004;34:364–372. doi: 10.1016/j.ymeth.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 61.Schulz G.E. A new classification of membrane protein crystals. J. Mol. Biol. 2011;407:640–646. doi: 10.1016/j.jmb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 62.Zaccai N.R., Serdyuk I.N., Zaccai J. Cambridge University Press; Cambridge, UK: 2017. Methods in Molecular Biophysics. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.