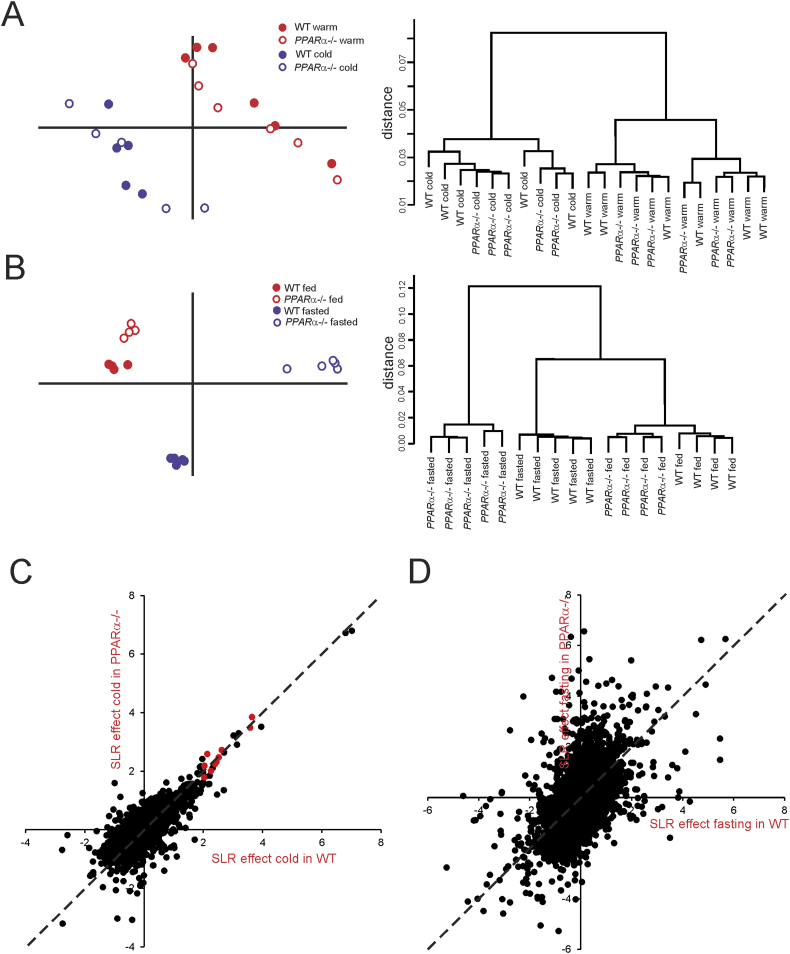
Figure 5.
Distinct clustering of inguinal WAT of thermoneutral and cold-exposed wildtype and PPARα−/− mice. (A) Left panel: Principle component analysis of transcriptomics data from inguinal WAT of the thermoneutral and cold-exposed wildtype and PPARα−/− mice. The graph shows the clear separation of mice according to treatment, while the two genotypes are mixed. Right panel: Hierarchical clustering of transcriptomics data from the thermoneutral and cold-exposed wildtype and PPARα−/− mice. The dendogram reveals the distinct clustering and separation based on treatment, but not based on genotype. (B) Left panel: Principle component analysis of transcriptomics data from livers of fed and fasted wildtype and PPARα−/− mice. The graph shows the clear separation between the genotypes in both fed and fasted state. Right panel: Hierarchical clustering of transcriptomics data from the fed and fasted wildtype and PPARα−/− mice. The dendogram reveals the distinct clustering and separation according to treatment and genotypes. (C) Correlation plot showing gene expression changes in response to cold in wildtype (x-axis) versus PPARα−/− mice (y-axis) (expressed as signal log ratio). Well established PPARα target genes are highlighted in red (Cox7a1, Slc27a2, Cidea, Gys2, Pank1, Gyk, Plin5, Pdk4, Slc25a20, Hadhb). (D) Correlation plot showing gene expression changes in response to fasting in wildtype (x-axis) versus PPARα−/− mice (y-axis) (expressed as signal log ratio).